Figures & data
Figure 1 The folate synthesis and metabolic pathways in M. tuberculosis and the target of para-aminosalicylic acid (PAS). MTHFR, 5,10-methylenetetrahydrofolate reductase; DHPS, dihydropteroate synthase; DHFS, dihydrofolate synthase; DHFR, dihydrofolate reductase; TS, thymidylate synthase; MetE, cobalamin-independent homocysteine transmethylase; MetH, cobalamin-dependent methionine synthase; HSMT, serine hydroxymethyltransferase.
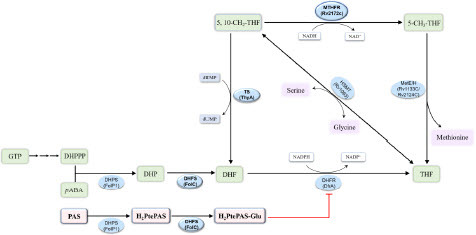
Figure 2 rv2172c T120P and M172 V mutations are enriched in the PAS-resistant (PAS-R) clinical isolates.
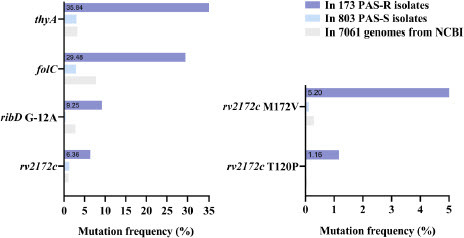
Table 1. rv2172c T120P or M172 V mutation leads to PAS resistance in M. tuberculosis
Figure 3 Predicting the effect of T120P and M172 V mutations on binding to nicotinamide adenine dinucleotide (NADH) based on the structure of MTHFR–coenzyme complex from different species. (A) Crystal structures of MTHFRs containing either NADH or flavin adenine dinucleotide (FAD) were visualized from a consistent perspective. FAD-dependent MTFHR: E. coli MetF (PDB ID: 1ZPT), N. meningitidis MetF (PDB ID: 7RML), H. influenzae MetF (PDB ID: 5UME), T. thermophilus TTHA_0327 (PDB ID: 3APY), S. cerevisiae MET12 (PDB ID: 6FNU). FAD-independent MTHFR: M. smegmatis MSMEG_6649 (PDB ID: 7WMZ). (B) Multiple sequence alignments of MTHFRs from different species. Residues potentially interacting with NADH or FAD are annotated. (C) Structure superimposition of Rv2172c (modeled by AlphaFold2) and MSMEG_6649–NADH complex (PDB ID: 7WMZ). Residues potentially interacting with NADH are listed in the embedded table. (D) Noncovalent interactions between Met172, Arg119, and Met121 with NADH in mycobacterium MTHFR.
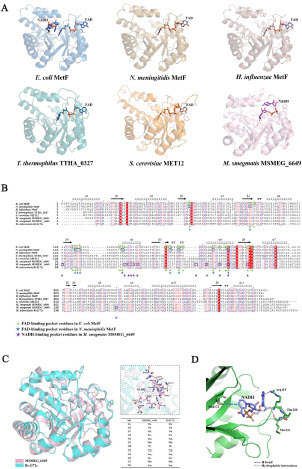
Figure 4 Enzymatic kinetics studies of Rv2172c, Rv2172c T120P, and Rv2172c M172 V. (A) Michaelis–Menten plots of proteins for NADH. Embedded table shows kinetic constants. (B) Michaelis–Menten plots of proteins for 5, 10-CH2-THF. Embedded table shows kinetic constants. The means ± standard deviations (SDs) were determined from two independent replicates.
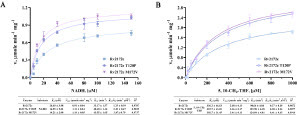
Figure 5 Relative MTHFR activities of Rv2172c, Rv2172c T120P, and Rv2172c M172 V. (A) Enzymatic activities are measured in reactions with 250 µM 5, 10-CH2-THF and varied NADH concentrations (10, 20, 40, and 80 µM) (Figure S1). (B) Enzymatic activities are measured in reactions with 100 µM NADH and varied 5, 10-CH2-THF concentrations (200, 400, 600, and 800 µM) (Figure S2). The relative enzymatic activities of the mutant proteins are normalized based on the enzymatic activity of RV2172c (presented as 100%) in each reaction. Data are presented as means ± SDs, in three independent experiments. **p < 0.01, ***p < 0.001.
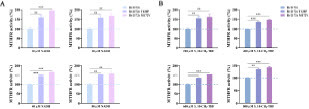
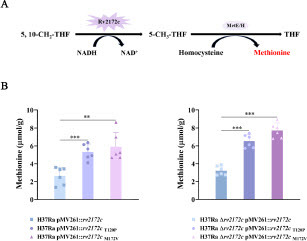
Figure 6 rv2172c T120P and M172 V mutations increase the biosynthesis of methionine in M. tuberculosis. (A) MTHFR-catalyzed reaction of Rv2172c in methionine synthesis pathway. (B) The intracellular methionine concentration in H37Ra pMV261::rv2172c, H37Ra pMV261::rv2172cT120P, H37Ra pMV261::rv2172cM172V, H37Ra Δrv2172c pMV261::rv2172c, H37Ra Δrv2172c pMV261::rv2172cT120P, and H37Ra Δrv2172c pMV261::rv2172cM172V strains. **p < 0.01, ***p < 0.001.