Figures & data
Table 1. Compounds of the scheme-1 (1–4 series) (N2-alkyl-N3-phenylquinoxaline-2,3-diamine derivatives): reaction time, yield, and M.P. of each compound
Scheme 1. Reagents and conditions: (a) (COOH)2.2H2O, 4 N HCl, 100°C, 6 h; (b) SOCl2, DCE, Cat. DMF, 100°C, 6 h; (c) Alkyl amine (derivatives), K2CO3, DMF, rt, 12 h; (d) Aniline (derivatives), ethanol, reflux, 12 h.
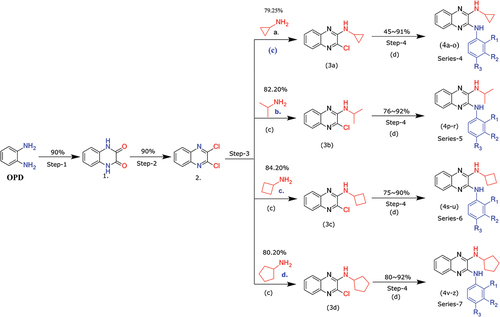
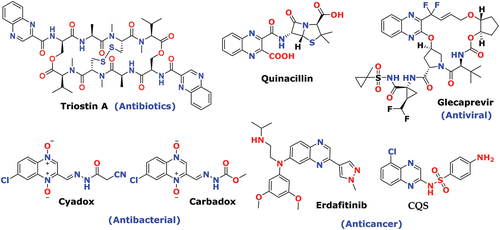
Figure 2. Novel designed from the privileged structure (a) Antibacterial activity from DNA intercalators [Citation66] (b) Hydrazine binding with specific amino acid of Dihydrofolate reductase protein. [Citation67].
![Figure 2. Novel designed from the privileged structure (a) Antibacterial activity from DNA intercalators [Citation66] (b) Hydrazine binding with specific amino acid of Dihydrofolate reductase protein. [Citation67].](/cms/asset/e8576be3-542c-42e7-bac6-47d4c37fea2d/teba_a_2049085_f0002_oc.jpg)
Table 2. Antimicrobial and antifungal activity, zone of inhibition (mm)
Table 3. Physicochemical property estimation (QikProp properties) of new designs
Table 4. Docking scores, residue contacts and ∆G of binding scores of proposed designs. Ch; Chloramphenicol
Figure 3. The comparison between the inhibition zones of compounds (4b-4z) and standard drugs against bacterial and fungal strains.
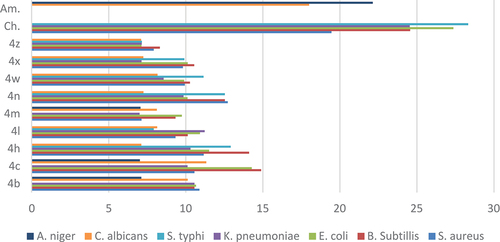
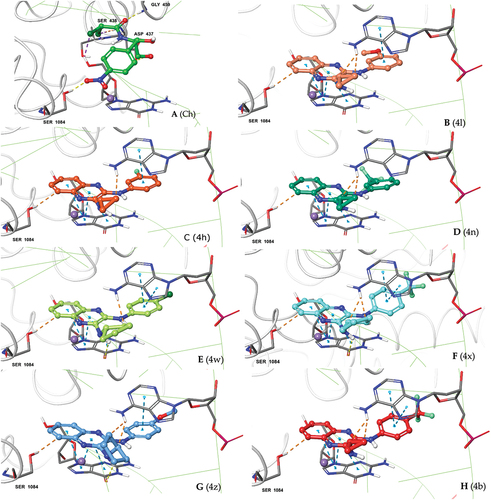
Figure 4. Docking poses of our 2,3-diamino quinoxaline designs (A-H) at the quinolone-binding site of S. aureus DNA gyrase. Consistency in binding interactions with protein (Ser-1084) and DNA base pairs DG (G):9; DA (H): 13 and posing adjucent to Mg+2 ion can be seen for all the new designs.