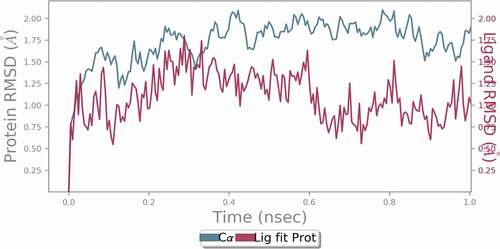
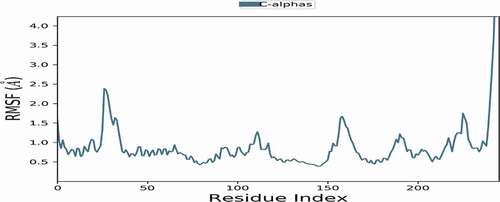
Figures & data
Table 1. Docking scores of the analyzed compounds against the 3ERT receptor with Tamoxifen as the reference
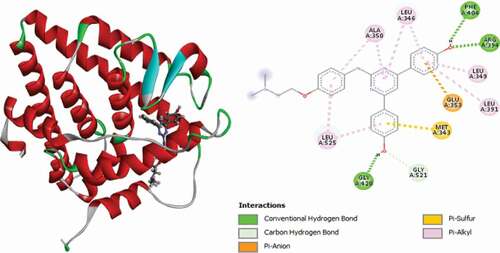
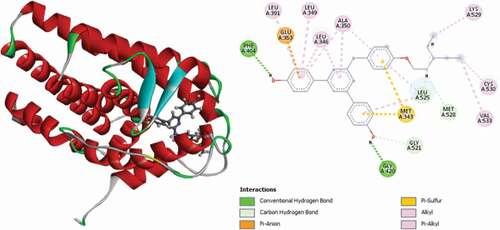
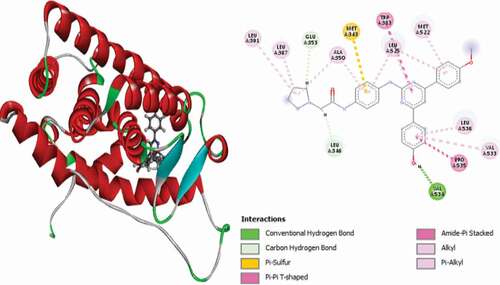
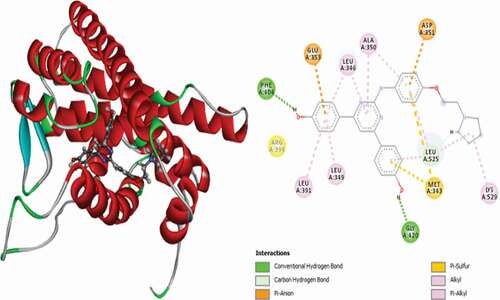
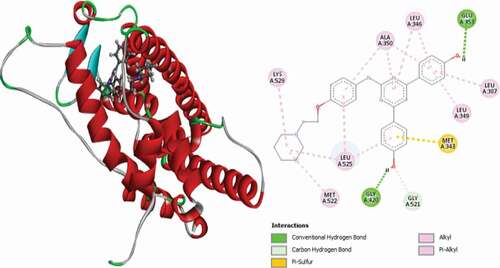
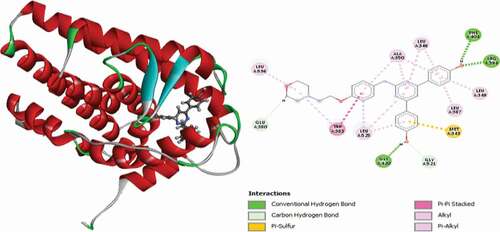
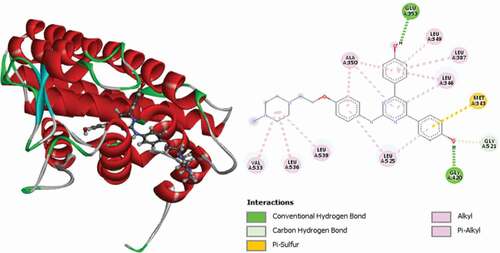
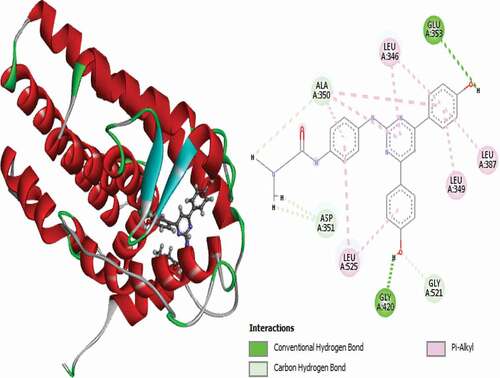
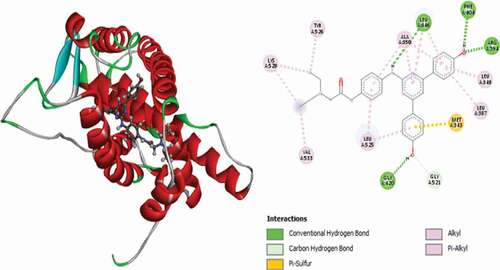
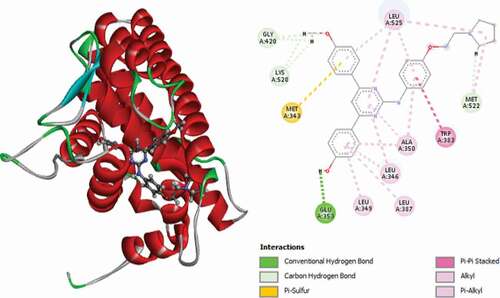
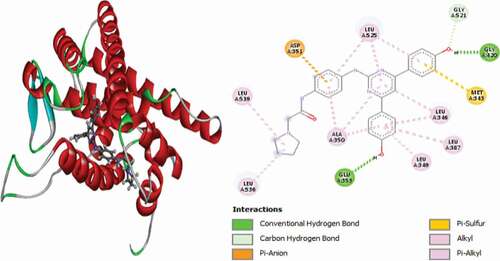
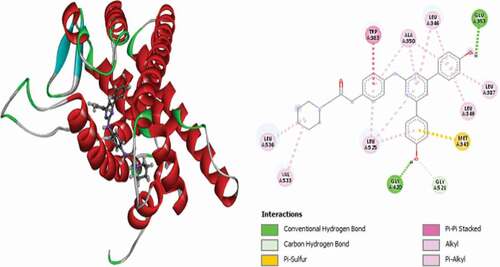
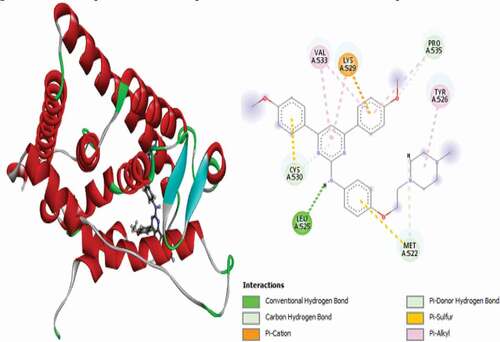
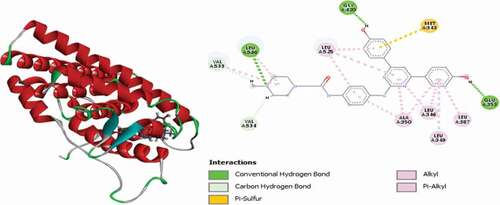
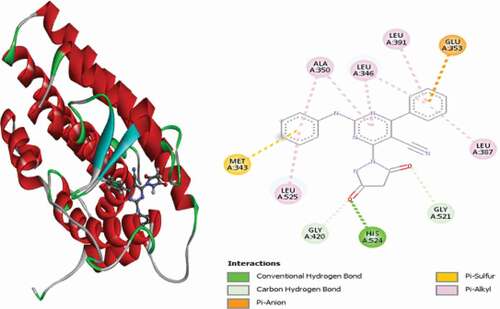
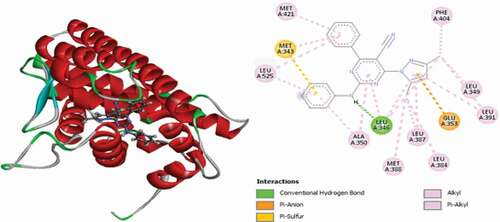
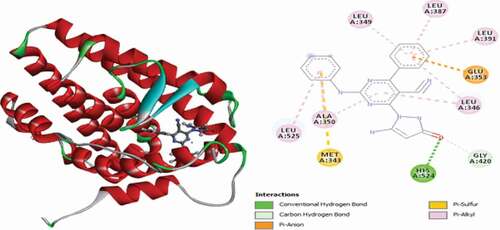
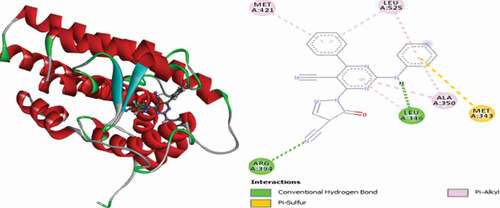
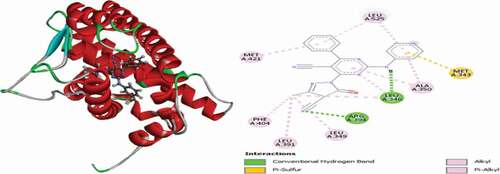
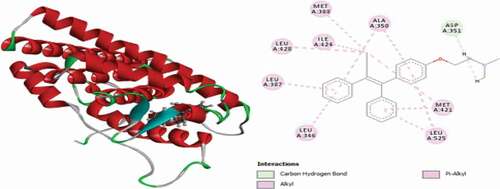
Table 2. Various interactions between the potential hit compounds and the active site of the 3ERT receptor
Table 3. Predicted drug-likeness properties of the selected compounds
Table 4. Predicted ADMET properties of the selected compounds