Figures & data
Table 1. Cervical cancer various stages
Table 2. Various researches that diagnose cervical cancer using machine learning approaches
Table 3. Description of the dataset
Figure 2. (a) Bar graph showing the percentage of null values in the dataset (b) The percentage of biopsy positive and biopsy negative results.
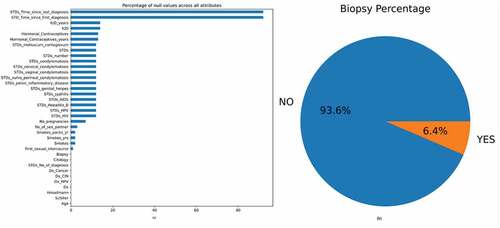
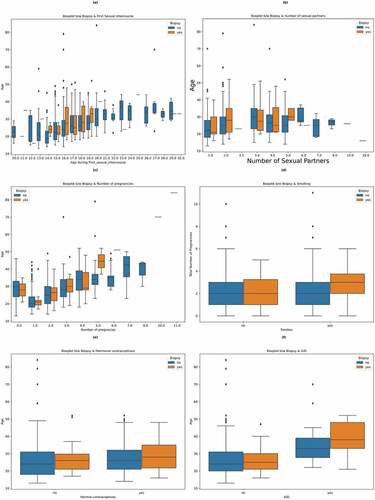
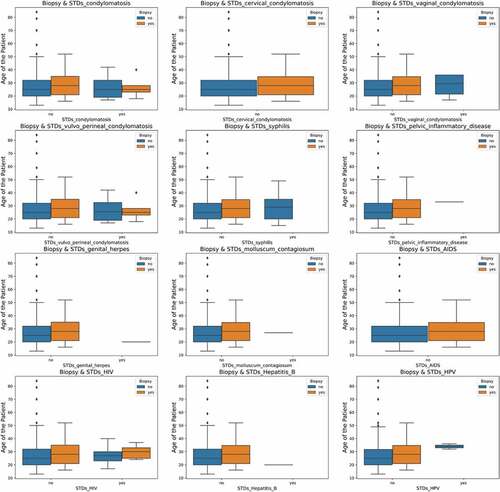
Figure 3. Count plots and density plots (a) The number of patients who smoke (b) The number of patients who use hormonal contraceptive (c) The number of patients who use intra-uterine devices (d) The number of patients who have sexually transmitted diseases (e) Patient age distribution (f) The number of pregnancies (g) The number of sexual partners.
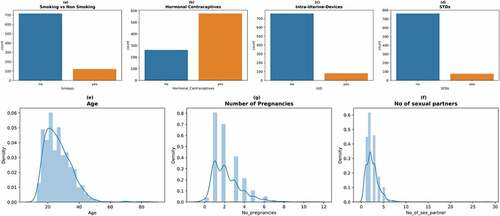
Figure 6. The presence of outliers in data. (a) Outliers before IQR treatment (b) After IQR treatment (Outliers removed).
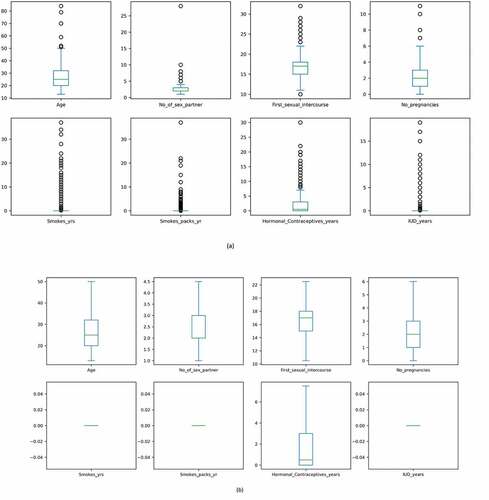
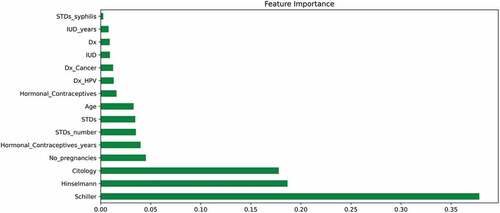
Figure 7. Mutual Information which describes the relationship among various attributes which diagnose cervical cancer.
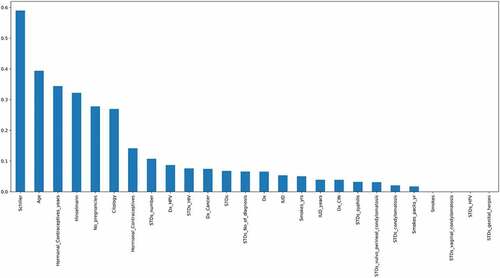
Figure 8. Pearson’s correlation heatmap which describes the relationship among various attributes which diagnose cervical cancer.
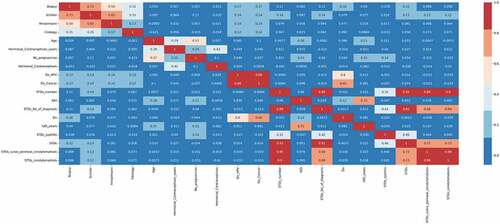
Figure 9. Biopsy results before and after balancing. (a) Initial unbalanced data (b) Balanced data after using Borderline-SMOTE.
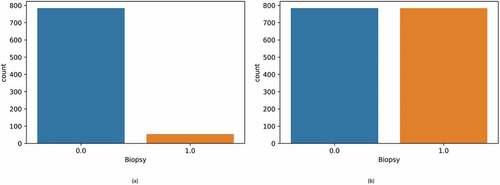
Table 4. Performance evaluation of classifiers without using feature selection and borderline-SMOTE (Unbalanced dataset)
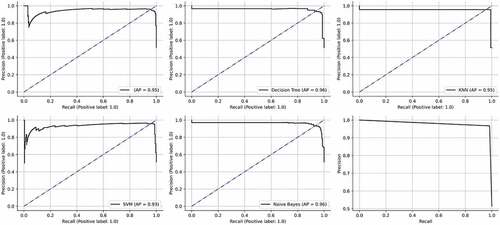
Figure 12. ROC curves obtained by the classifiers. (a) Logistic regression (b) Decision tree (c) KNN (d) SVM (e) Naïve Bayes (f) STACK A.
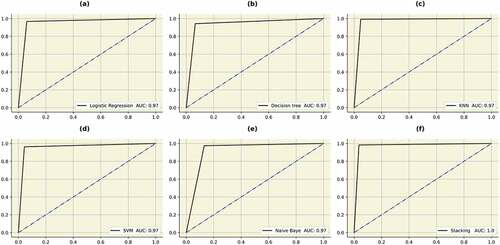
Table 5. Performance evaluation of the initial set of classifiers after data balancing and hyperparameter tuning
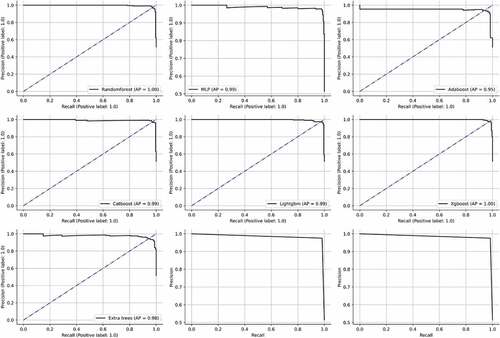
Figure 14. AUC’s of the bagging, boosting and stacking classifiers. (a) Random forest (b) MLP (c) Adaboost (d) Catboost (e) Lightgbm (f) Xgboost (g) Extratrees (h)STACKB (i) STACKC.
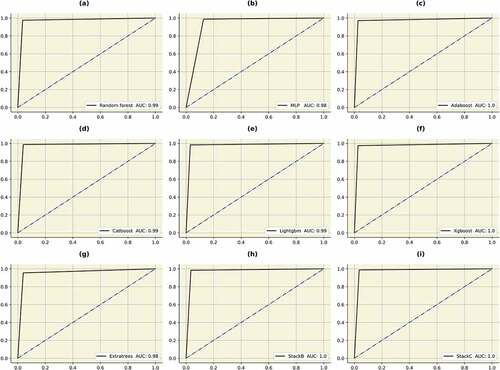
Table 6. Performance evaluation of bagging and boosting classifiers after data balancing and hyperparameter tuning
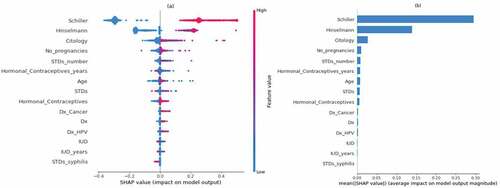
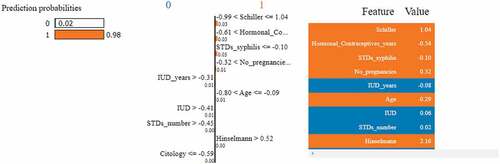
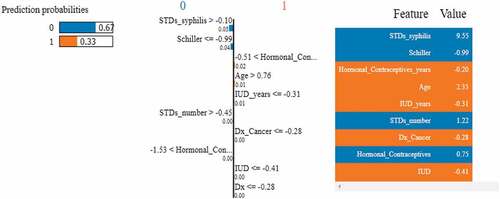
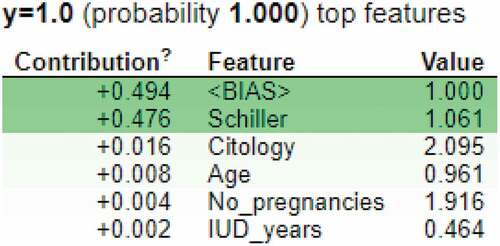
Figure 21. Feature importance using LIME graphs. (a) Positive biopsy result (b) negative biopsy result.
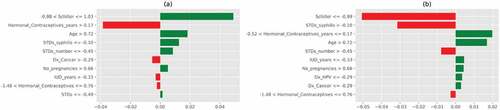
Figure 23. User interface of the prediction model deployed using “Gradio” to classify cervical cancer results.
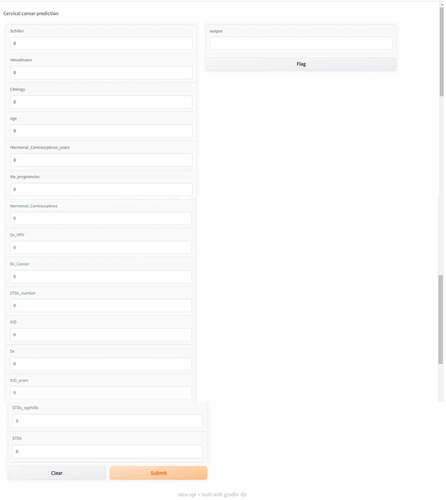
Table 7. Comparison of various researches