Figures & data
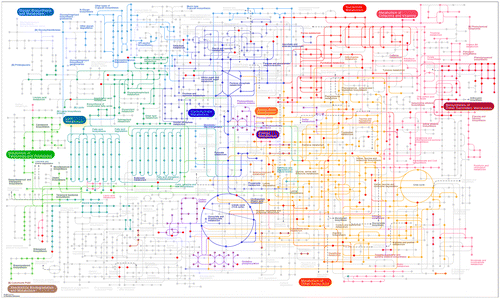
Figure 1. Groups of metabolites and their possible relatedness in B. napus seeds as inferred from STITCH 5.0 (Szklarczyk et al., Citation2016) small molecule database mining of total 160 metabolites of which 110 could be assigned to KEGG IDs. For the compound list please refer to Supplementary Table 1.
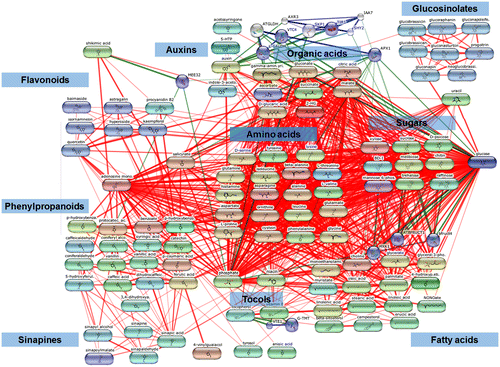
Figure 2. KEGG metabolic pathway showing the mapped 110 metabolites (highlighted as bold red dots) obtained from B. napus seeds through metabolomics approaches.