Figures & data
Figure 1. RaFoSA improves visualization of molecules when only coarse-grained information is available. (a) 1W0M, and (b) 2AGV are protein molecules arranged in an apparent order of increasing SS complexities. The visualizations on the left side (when all-atom and SS information are lacking) are not helpful and do not allow one to clearly differentiate (more structurally stable) sheets from (less structurally stable) coils. In contrast, the visualizations on the right side when RaFoSA provides SS information are more useful/helpful and are visually appealing. With the SS assigned, one can see the residues that form more structurally stable sheets (red) and helixes (blue), or less structurally stable coils (black).
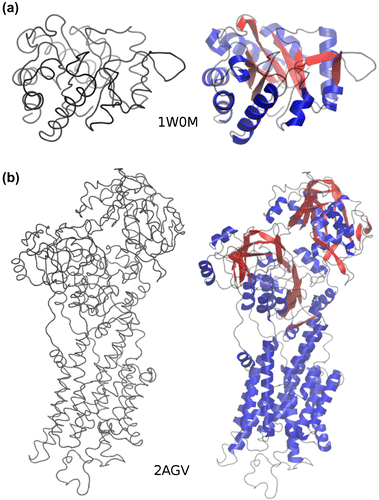
Table 1. Agreements between various SS assignment methods and DSSP (Kabsch & Sander, Citation1983) and STRIDE (Heinig & Frishman, Citation2004)
Figure 2. Features used in RaFoSA. One of the features is the residue type (which is any of the 20 standard amino acids or “X” for any non-standard amino acid). Other features are related to alpha carbon (CA) atoms. Six of the features are CA-CA distances (a), such as di−1,i+1. Other six of the features are CA-CA-CA angles (b), such as ai−2,i,i+2. Four of the features are sign or angle of CA-CA-CA-CA torsional angles (c), such as ti−1,i,i+1,i+2. While the remaining features are based on residue–residue contacts (c), such as Ci,4.0. “i” is the index of the current residue. “i − 1” (or “i + 1”) is the index of the residue immediately before (or immediately after) the current residue.
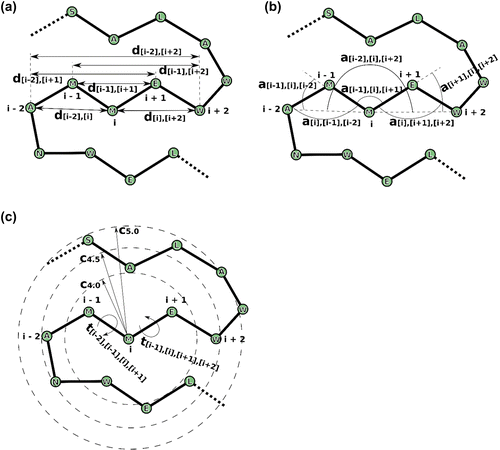
Figure. 3. Agreement between RaFoSA’s SS assignment and SS assignment by some existing methods. (a) Proportion of the assigned SS that falls in each of the seven SS classes–alpha helix (H), beta sheet (B), strand (E), 3–10 helix (G), pi-helix (I), turn (T), and coil/bend (S). (b) Proportion of the assigned SS that falls within each of the three SS classes–sheet, s; helix, h; coil, c–based on mapping 1, M1 (“HBEGITS” → “hcscccc”). (c) Same as b, but based on mapping 2, M2 (“HBEGITS” → “hsshhcc”). Panels d to f (based on M1) and panels g to h (based on M2) show the degree of agreement between RaFoSA and each of the other methods. Using M1, agreements between SS assignments by RaFoSA and DSSP are shown in d, between RaFoSA and STRIDE are shown in e, and between RaFoSA and PSEA are shown in f. Similar information are shown in g, h, and i, but based on M2. The columns in each of the matrixes are for RaFoSA-assigned Sheets, Helixes, and Coils, respectively. The rows are for the other SS methods RaFoSA is compared to. The intensity of the blue color in the leading diagonal of each of the matrixes show the degree of agreement between RaFoSA and the other SS assignment method. Number of amino acids per SS are shown in j (sheet), k (helix), and l (coil).
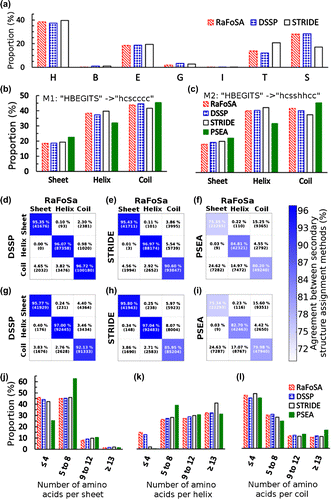
Figure. 4. Structural stability score based on RaFoSA-assigned SS identifies protein’s structurally stable variant. Structural stability of wild type B. subtilis LipA (blue, 2QXU), and its variant X mutant (red, 3QZU) over the simulation time at three different temperatures—(a) 300 K, (b) 350 K, and (c) 400 K—are shown. The solid lines are the means, while the dotted/dashed lines are one standard deviation above or below the respective mean values.
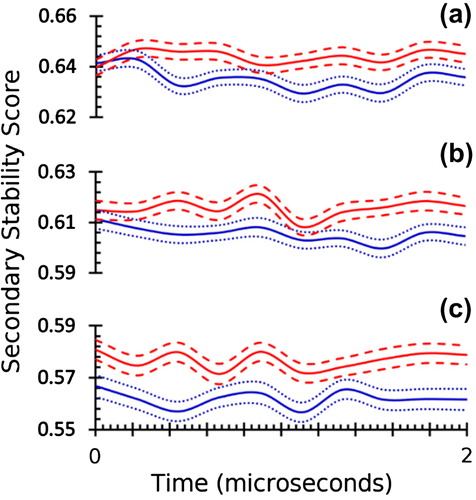
Figure. 5. RaFoSA webserver. The webserver accepts input (a) in any of four ways: (1) PDB file, (2) PDB content/format as text, (3) PDB ID, or (4) trajectory as PSF and DCD files. SS are assigned for all frames (b) in the submitted data, and SS visualization (c) is generated for the protein for each frame. Summary statistics (line graph, (b), and doughnut chart, (c)) are also provided for the assigned SS.

