Figures & data
Table 1. The average day length and water temperature in four seasons from where fish were collected (latitude 24°44′N, longitude 93°58′E)
Table 2. Details about primer sequences used for RT-qPCR for Tph1, Aanat1, Aanat2, and Hiomt in the brain and gut of Catla catla
Figure 1. Seasonal and light–dark phase changes in the expression level of (A) Tph1 in Brain, (B) Tph1 in Gut and (C) Aanat1 in Brain, (D) Aanat1 in Gut were compared by taking the mean value of three time points during the light phase (represented by empty circle) and the dark phase (represented by filled circle).
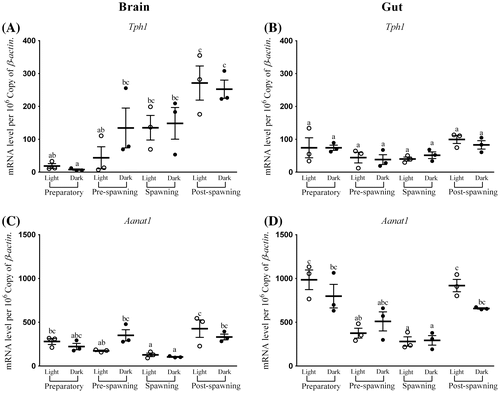
Figure 2. Seasonal and light–dark phase changes in the expression level of (A) Aanat2 in Brain, (B) Aanat2 in Gut and (C) Hiomt in Brain, (D) Hiomt in Gut were compared by taking the mean value of three time points during the light phase (represented by empty circle) and the dark phase (represented by filled circle).
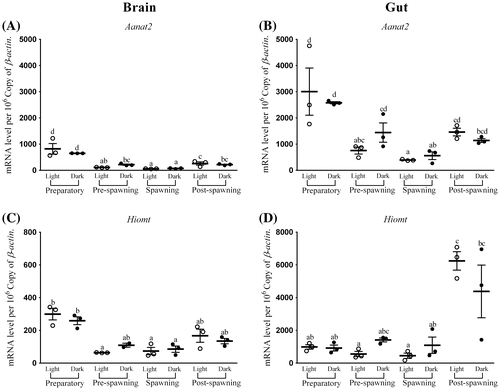
Figure 3. Changes in the expression of Tph1 in six different time points (8, 12, 16, 20, 24, and 4 h) in different seasons (n = 3) were fitted to nonlinear regression curve using the formula “Y = Mesor + Amplitude Cos (Frequency X + Acrophase)”.
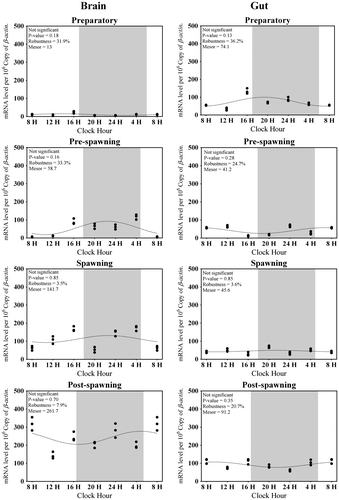
Table 3. Correlations study between water temperature and expression level
Figure 4. Changes in the expression of Aanat1 in six different time points (8, 12, 16, 20, 24, and 4 h) in different seasons (n = 3) were fitted to nonlinear regression curve using the formula “Y = Mesor + Amplitude Cos (Frequency X + Acrophase)”.
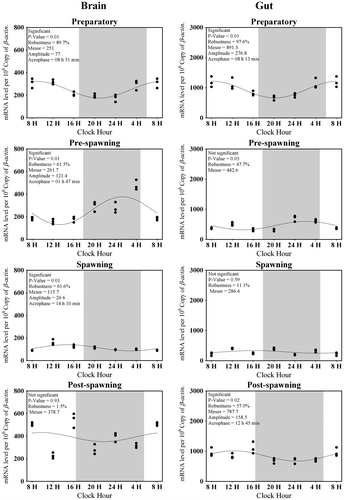
Figure 5. Changes in the expression of Aanat2 in six different time points (8, 12, 16, 20, 24, and 4 h) in different seasons (n = 3) were fitted to nonlinear regression curve using the formula “Y = Mesor + Amplitude Cos (Frequency X + Acrophase)”.
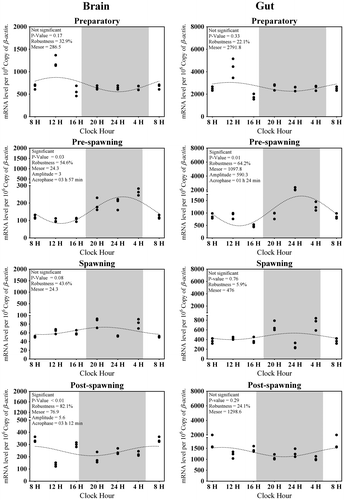
Figure 6. Changes in the expression of Hiomt in six different time points (8, 12, 16, 20, 24, and 4 h) in different seasons (n = 3) were fitted to nonlinear regression curve using the formula “Y = Mesor + Amplitude Cos (Frequency X + Acrophase)”.
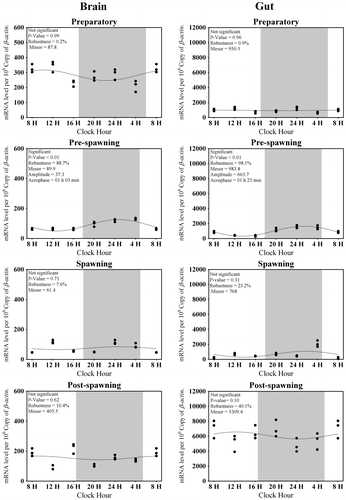
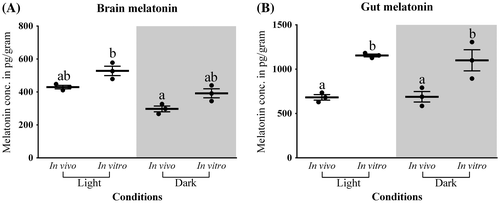
Figure 7. In vivo and in vitro melatonin synthesis in (A) Brain and (B) Gut tissues was analyzed by two-way analysis of variance followed by Student–Newman–Keuls.