Figures & data
Figure 1. Distribution of IAV subtypes isolated from gulls.
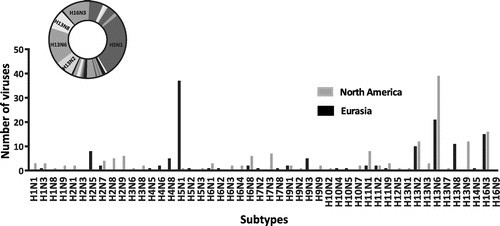
Figure 2. Time-clock Bayesian inference analysis of H13 nucleotide sequences from North America and Eurasia.
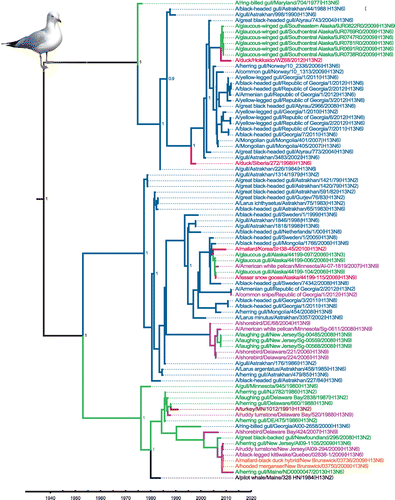
Figure 3. Time-clock Bayesian inference analysis of H16 nucleotide sequences from North America and Eurasia.
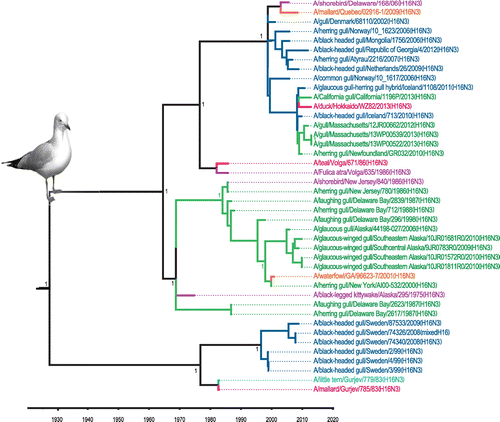
Figure 4. Putative geographic progression of gull virus genes that contributed to the genesis of the Eurasian H16N3 gull virus identified in Newfoundland, Canada.
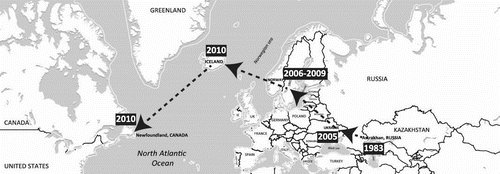
Table 1. Experimental infections of gulls with avian strains
Table 2. Experimental infections of avian and mammalian hosts with gull strains
Table 3. Gull virus binding assays on avian and human tissues (Lindskog et al., Citation2013)
Figure 5. Phylogenetic analysis of gull H5 and H7 nucleotide sequences.

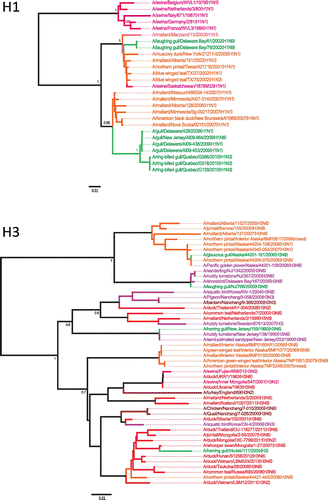
Figure 6. Phylogenetic analysis of gull H1 and H3 nucleotide sequences.