Figures & data
Table 1. Grade description for screening against M. oryzae
Table 2. Chromosome location (CL), size range (SR), primers sequence and annealing temperature (AT) for two SSR markers used in the study
Table 3. Leaf blast severity were recorded by following 0–9 SES scale as per IRRI, Philippines
Figure 1. Activity profile of superoxide dismutase (SOD).
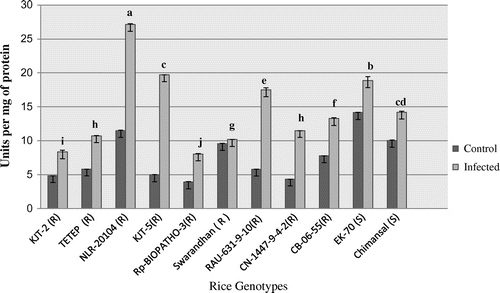
Figure 2. Activity profile of peroxidase (POX).
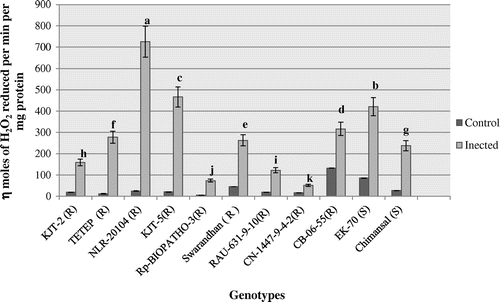
Figure 3. Activity profile of ascorbate peroxidase (APX).
Figure 4. Activity of phenylalanine ammonia lyase (PAL).
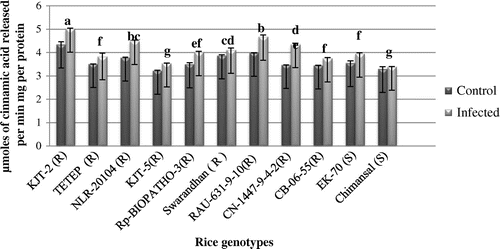
Figure 5. Activity of Chitinase.
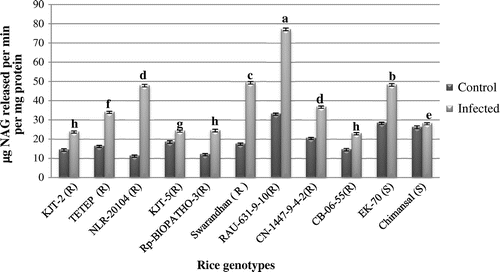
Figure 6. Activity of β-glycosidase.
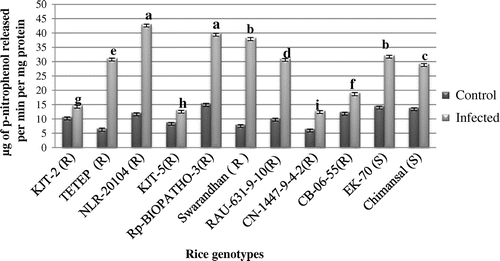
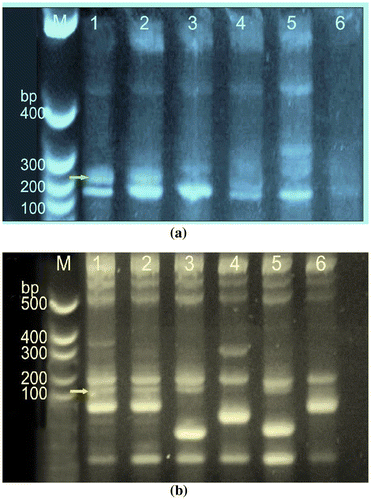
Figure 7. The purified genomic DNA was quantified and equal amount of DNA was used for PCR amplification. (a) Amplified genomic DNA of blast resistant and susceptible genotypes with SSR primer RM 224 and (b) with SSR RM144. The Lane M:100 bp, lane 1:1-KJT-2 (R),Lane 2:KJT-5 (R), Lane 3-TETEP (R), Lane 4:Chimansal(S), Lane 5:NLR-20104 (R),Lane 6:-EK-70 (S).