Figures & data
Figure 2. Scanning the Gata3jal allele for an integrated intracisternal A particle (IAP) element. (a) Annealing sites for a gene-specific primer (GSP) pair, one in the forward and one in the reverse orientation, are shown within an intron of the Gata3 gene (represented by a green line with the sense orientation indicated). In the top diagram, the GSP are close enough together to yield a PCR product of the wild-type length. In the bottom diagram, an IAP (represented by a thick blue line) has integrated between the primer-annealing sites, moving the GSP pair so far apart that no product is generated in the PCR. (b) One GSP pair, designated GSP-F and GSP-R (sequences given in Figure ), produced a 1,016 bp amplimer from wild-type DNA templates, but no products from jal/jal mutant DNA templates, as expected if an IAP integration occurred between the primer-annealing sites. (c) Here an intron with an IAP insertion is depicted in each of the two possible orientations, sense (top diagram) or antisense (bottom diagram), with respect to the Gata3 gene. If the IAP is integrated in the sense orientation, GSP-F should yield a PCR product with IAP-specific primer (ISP) -R1, and GSP-R should yield a product with ISP-F2. If the IAP is integrated in the antisense orientation, GSP-F should yield a PCR product with ISP-R3, and GSP-R should yield a product with ISP-F4. (d) GSP-F yielded a 959 bp product from jal templates with ISP-R3 but none with ISP-R1. GSP-R yielded a 486 bp product from jal templates with ISP-F4 but none with ISP-F2. Therefore, the IAP integrated in Intron 3–4 in the Gata3jal allele is in the antisense orientation with respect to the gene.
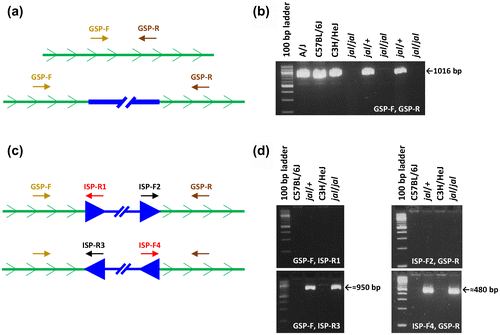
Figure 3. Nucleotide sequence at the site of IAP integration in Intron 3–4 of the mutant Gata3jal allele. Sequences in lower case are located within Gata3, Intron 3–4 (beginning with nucleotide number 2:9869836); nucleotides from Exon 4 are shown in blue, upper case letters (ending with nucleotide number 2:9868821). Sequences that correspond to the primers described in the text and in Figure as GSP-F and GSP-R are highlighted in yellow. The six-nucleotide direct repeat created in mouse genomic DNA by IAP element insertion (the target site duplication, TSD) is shown in red (and corresponds to nucleotides 2:9869167–9869162). The IAP element’s 5’ and 3’ LTRs, in antisense orientation, are shown in green; IAP sequences located between the LTRs are abbreviated by blue stars. Sequences that correspond to IAP-specific primers described in the text and in Figure as ISP-R3 and ISP-F4 are highlighted in blue and pink, respectively. Base-pair positions on mouse Chromosome 2 are from NCBI Build 37.2 (Ensembl Mouse Genome Server, Citation2016; Mouse Genome Database, Citation2016).

Figure 4. Gata3IAP and jal are meiotically inseparable among a panel of 374 backcross mice. (a) Ramirez et al. (Citation2013) described the production of a panel of 374 backcross mice from a cross of (A/J x C3H/HeJ-jal/J)F1 females with C3H/HeJ-jal/J mutant males. These panel members segregated for jal and for various other markers on proximal Chromosome 2. Here, the most salient 10 members of that panel (i.e., those with crossovers nearest the jal mutation) are each represented by a line that depicts the region of Chr 2 between markers SNP1 (officially designated rs27112885) and SNP3 (a.k.a. rs27100936) that was inherited from their F1 parent. A/J-derived sequences are drawn in black, and C3H-derived sequences are drawn in red. Each recombinant is identified by its pedigree number, followed by a letter to indicate its phenotype as wild type (w) or mutant (j). Six crossovers (found in recombinants 62, 92, 328, 343, 44, and 211) lie telomeric to SNP1 but centromeric to jal; one crossover (found in recombinant 309) lies telomeric to jal but centromeric to SNP2 (a.k.a. rs27131571). Thus, jal must lie between SNP1 and SNP2, an interval that includes only one gene known to be expressed in skin, Gata3. Official SNP designations are from dbSNP Build 142 (Ensembl Mouse Genome Server, Citation2016; Mouse Genome Database, Citation2016). (b) Recombinant DNA samples from the panel described by Ramirez et al. (Citation2013) and controls were characterized for the presence of none, one, or two copies of Gata3IAP. All recombinants that were phenotypically mutant were homozygous for Gata3IAP, and the wild-type recombinants, heterozygous jal/+, were heterozygous for Gata3IAP/Gata3+. Co-segregation of Gata3IAP and jal in this backcross suggests that they are very close or identical.
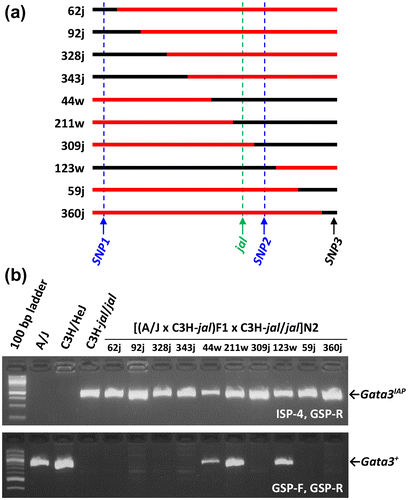
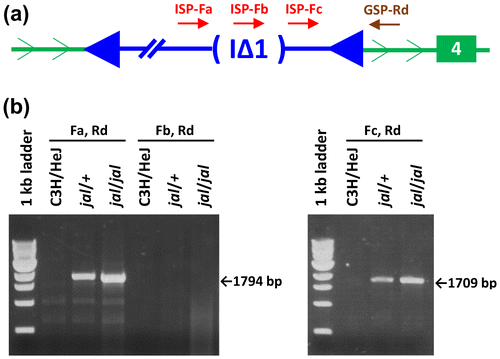
Figure 5. Characterization of the IAP subtype associated with the Gata3jal insertion. (a) The relative location of the ISP-Fa, -Fb, -Fc and GSP-Rd annealing sites are shown. Note that each of these primer sequences is displayed in Supplementary Figure S1. (b) The ISP-Fa, GSP-Rd primer pair should produce a jal-specific amplimer only if the 1.9 kb gag-pol deletion characteristic of IAP elements of the I∆1 subtype should position primer-annealing sites for -Fa and -Rd near one another, similar to the ISP-Fc to GSP-Rd distance. The I∆1 deletion should remove the annealing site for ISP-Fb, so a deleted IAP template is not expected to produce an amplimer with the -Fb, -Rd pair. The -Fa and -Rd pair directed the amplification of a 1,794 bp product from jal templates, only 85 bp longer than the 1,709 bp jal-specific product yielded by the -Fc, -Rd primer pair, suggesting that the Gata3jal-associated IAP is of the I∆1 subtype. The full DNA sequence of the jal-specific, -Fa to -Rd amplimer (which includes the I∆1 deletion breakpoints in the Gata3jal-associated IAP insertion) is shown in Supplementary Figure S1.