Figures & data
Figure 1. Construction of pET-GST-YC1 expression vector. (A) The composition of the pET-GST-YC1 vector. (B) The amino acid sequences of ProTX II, GsAF II, and YC1.
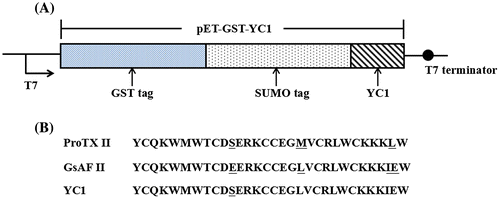
Figure 2. Expression of GST-YC1 in SHuffleTM.
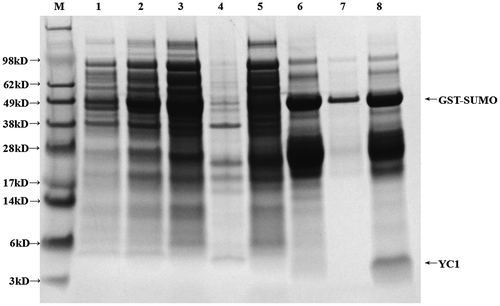
Figure 3. RP-HPLC chromatography of rYC1. (A) Absorption of the eluted proteins was measured at 280 nm, and fractions corresponding to detected peaks were collected. (B) Absorption of the eluting solution was measured at 215 nm, and fractions corresponding to detected peaks were collected.
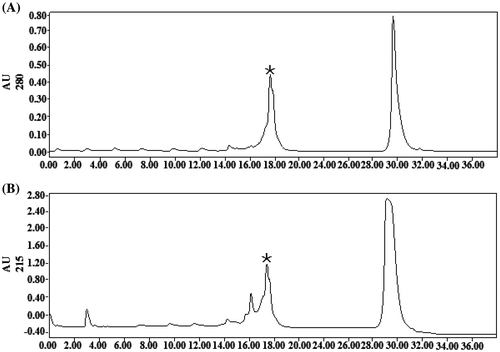
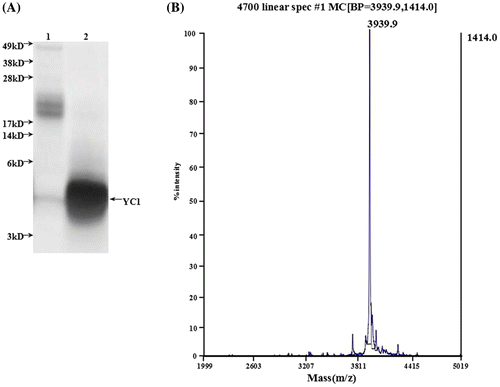
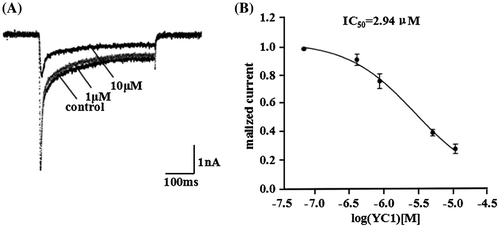
Figure 4. Identification of rYC1. (A) The results of the glycine-SDS-PAGE analysis. Lane 1, protein from eluent marked with asterisk in Figure ; lane 2, the protein after the ultrafiltration purification of eluent marked with asterisk (Figure ). (B) Mass spectra of the rYC1. The calculated theoretical molecular weight of rYC1 was 3,937.7 Da, and the measured molecular weight was 3,939.9 Da,