Figures & data
Figure 1. Annotated schematic representation of the mitochondrial genome of Betadevario ramachandrani. GenBank accession number MH817023.
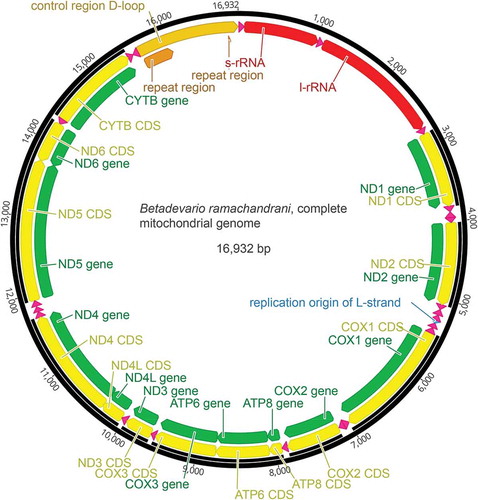
Figure 2. Phylogram of all species of subtribe Danionina represented on GenBank on the 17 May 2018, based on Bayesian analysis of nearly complete mitochondial genome sequences, with Sundadanio rubellus as outgroup. All nodes have Bayesian posterior probability = 1. Terminal labels end with GenBank accession number. Branch lengths are proportional to number of expected substitutions per site.
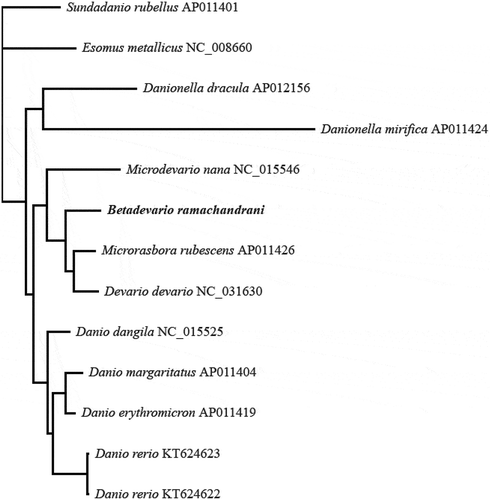
Figure 3. Two example chimeric genomes aligned to a genome of the main contaminant (Ctenopharyngodon idella). The numbers at top indicate position in the aligned genomes. The top bar indicates identity: if both sequences are identical, the bar is tall and bright green. As the aligned sequences come from distantly related taxa, they should not have significant identical regions, so bright green effectively indicates contamination. The grey bars are the aligned sequences, with dissimilar bases highlighted in black. The bottom bar indicates gene location, with arrows pointing in read direction. (A): KP407138 (“Devario chrysotaeniatus”) is nearly identical to C. idella, with the exception of one region spanning parts of the COI, COII and ATP6 genes, and one region spanning parts of the 12S and 16S rRNA genes; the first region is 99% similar to Procypris mera, a contamination, and the second is 97% similar to Devario laoensis, and possibly correct. (B): Contamination from C. idella in NC_028526 (“Danio margaritatus”) is evident in the ND5 gene, and in short regions flanking protein coding genes.

