Figures & data
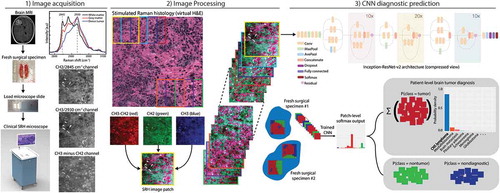
Figure 1. The SRH-CNN pipeline for automated intraoperative brain tumor diagnosis. (1) A patient newly diagnosed with a brain lesion undergoes a brain biopsy or planned resection. Fresh tissue is loaded directly into a stimulated Raman histology (SRH) imager for image acquisition. Images are acquired at two Raman shifts, 2,845 cm−1 and 2,930 cm−1, and a third image channel is generated via pixel-wise subtraction. Time to acquire a 1 × 1-mm2 SRH image is approximately 2 min. (2) A dense sliding window algorithm generates image patches that are preprocessed to optimize image contrast. (3) Each patch undergoes a feedforward pass through the network. Our inference algorithm is designed to retain the patches with high probability of being diagnostic, filtering the regions that are normal or nondiagnostic. Patch-level predictions from tumor regions are then summed and renormalized to generate a patient-level probability distribution over the diagnostic classes. Our pipeline can provide tissue diagnoses in <2.5 min using a 1 × 1-mm2 image, decreasing time to diagnosis by a factor of 10 compared with conventional intraoperative histology.
MRI, magnetic resonance imaging; H&E, hematoxylin and eosin; CNN, convolutional neural network.