Figures & data
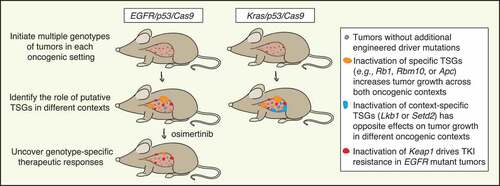
Figure 1. Dissecting the role of tumor suppressor genes using multiplexed genome editing across oncogenic contexts. Schematic of multiplexed genome editing in a mouse model of Epidermal growth factor receptor with activating point mutation L858R (EGFRL858R) mutant and Transformation related protein 53 (Trp53, best known as p53)-deficient lung adenocarcinoma (expressing Cas9; EGFR/p53/Cas9). Tumors are initiated by intratracheal administration of a lentiviral pool of vectors that lead to simultaneous tumor suppressor gene (TSG) inactivation mediated by CRISPR/Cas9 system. We modeled EGFR/p53 mutant tumors and 10 tumor genotypes via inactivation of putative tumor suppressor genes and quantified their impact on tumor growth compared to a different oncogenic context (Kirsten rat sarcoma viral oncogene homologue with activating point mutation G12D, KrasG12D mutant and p53-deficient model; Kras/p53/Cas9) and on sensitivity to treatment with osimertinib.