Figures & data
Figure 1. Pan-cancer and cancer type-specific aneuploidy patterns a. Frequency of chromosome arm aneuploidies from most to least common across all TCGA cancer types. The frequencies of cancer type-specific aneuploidy patterns highlighted in b are depicted in light blue and light red. The directionality of aneuploidy patterns (gain vs. loss) for chromosome arms 20q and 17p are highlighted. 20q is the most commonly gained and most infrequently lost chromosome arm, whereas 17p is the most frequently lost chromosome arm and only rarely gained. Bars for chromosome arm gains are red and losses are blue. Frequencies of chromosome arm aneuploidies are taken from reference.Citation13 b. Heat map of the average copy number data for somatic chromosome arms of BRCA (Breast Invasive Carcinoma), COAD (Colon Adenocarcinoma), and GBM (Glioblastoma Multiforme) cancers from the TCGA data set. Copy number gains are red and copy number losses are blue. Frequencies are taken from reference.Citation8 Characteristic cancer type-specific aneuploid chromosomes are surrounded by a black square.
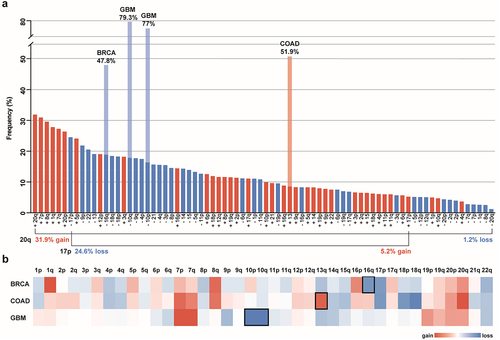
Figure 2. Aneuploidy patterns have been identified in all stages of cancer. Schematic of identified aneuploidy patterns in cancer or cell lines in the context of cancer evolution. The gray boxes display examples of recurring aneuploidy patterns. Boxes with dashed lines represent correlative evidence; boxes with solid lines represent direct evidence based on cell culture experiments. GBM: Glioblastoma Multiforme, ccRCC: clear cell Renal Cell Carcinoma, FHWT: Favorable Histology Wilms Tumor, BRCA: Breast Invasive Carcinoma, HCC: Hepatocellular Carcinoma, COAD: Colon Adenocarcinoma, HNC: Head and Neck Cancer, NSCLC: Non-Small Cell Lung Cancer.
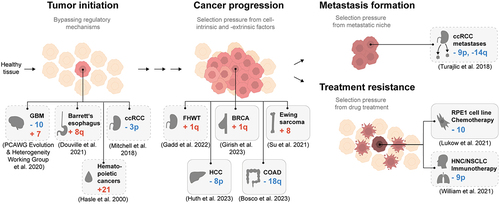
Table 1. List of putative driver genes of aneuploidy patterns for the 10 most common pan-cancer chromosome arm aneuploidies. Hallmark OGs and TSGs from the Catalogue of Somatic Mutations in Cancer (COSMIC) database with an aneuploidy frequency above 25% in the indicated tumor type.
Figure 3. Selection forces underlying the formation of aneuploidy patterns in cancer and human cell lines. Illustration of selection forces that shape the formation of aneuploidy patterns based on the observations from cancers, human cell lines, and model systems. Depending on environmental factors, cell type, and chromosome identity, aneuploidy patterns can be driven by positive, negative, or neutral selection forces. Positive selection forces underlying the copy number changes of a chromosome arm include: drivers of increased proliferation (upper left panel), compensating for harmful alterations (middle left panel) and conferring drug resistance (lower left panel). Selection forces that could be positive or negative depending on the context include: copy number loss resulting in loss of hetero-zygosity (LOH) and genetic interactions between aneuploid chromosomes (upper middle panels). Negative selection forces against chromosome copy number changes include: antagonization of proliferative alterations (upper right panel), the general proliferation decrease resulting from the imbalanced expression of many genes (middle right panel), and triplosensitive or haploinsufficient genes (lower right panel). Neutral selection of aneuploidy patterns is likely very rare and might only occur for aneuploidy of the sex chromosomes in specific cancer or cell types (lower middle panel). Chromosome arm gain is depicted in red, chromosome arm loss in blue.