Figures & data
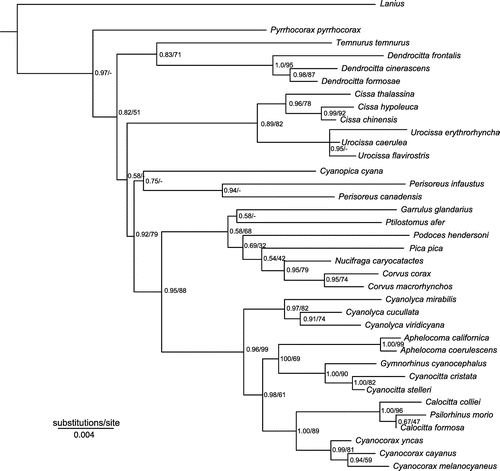
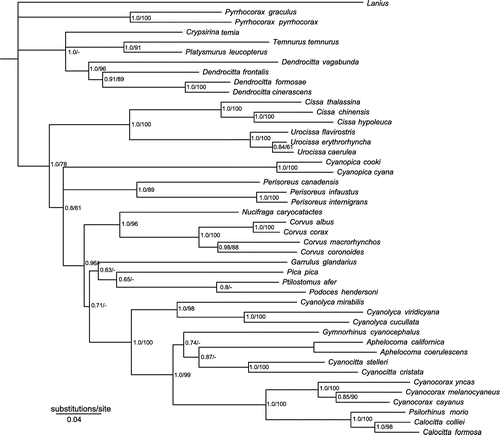
Figure 1. Phylogeny of Corvidae based on Bayesian and Maximum Likelihood analyses of the combined data-set. Numbers at the nodes refer to Bayesian posterior probability/ML bootstrap support. Asterisks indicate 1.0 posterior probability and 100% bootstrap support. Letters at nodes indicate five major clades of the family.
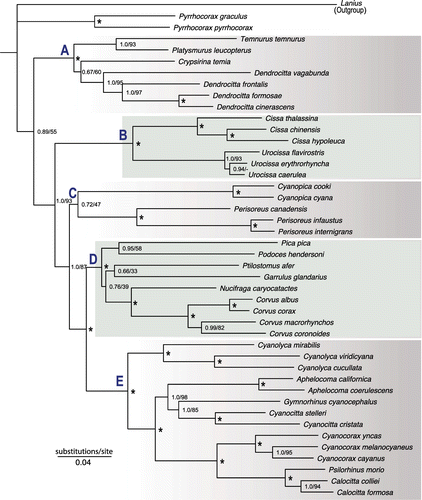
Figure 2. Reconstruction of the ancestral area of lineages of Corvidae. Pie charts indicate probability of various ancestral area combinations. Pie charts at nodes were derived from S-DEC analysis; the smaller charts above and below at the nodes obtained from S-DIVA and BBM analyses, respectively. Ancestral areas are shown only for basal nodes. The New World jay clade (clade E), its sister clade (clade D) and all other basal clades (C, B and A) of corvids, are outlined in light blue, orange and green boxes, respectively.
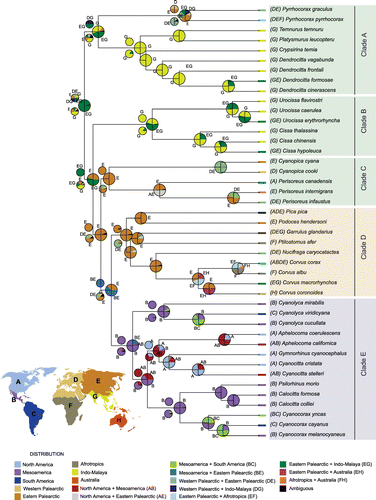
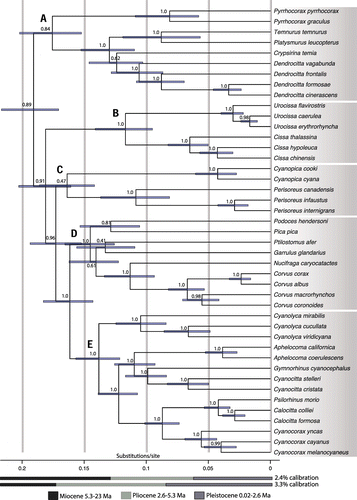
Figure 3. Approximate timeframe for the diversification of the Corvidae, with particular focus on New World jays. Time scales are based on ND2 substitution rates of 2.4% and 3.3% per lineage per million years. Bars at nodes indicate the highest posterior density (HPD) interval. Numbers at nodes refer to Bayesian posterior probabilities. Letters at nodes indicate the five major clades of the family.