Figures & data
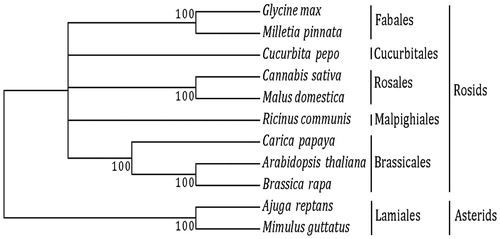
Figure 1. Maximum likelihood tree. We used 17-shared genes from nine species from the subclass Rosids: Order Brassicales – Arabidopsis thaliana (NC_001284), Brassica rapa (NC_016125), and Carica papaya (NC_012116); Order Cucurbitales – Cucurbita pepo (NC_014050); Order Fabales – Glycine max (NC_020455), and Millettia pinnata (NC_016742); Order Malpighiales – Ricinus communis (NC_015141); and Order Rosales -–Malus domestica (NC_018554), and Cannabis sativa (KR_059940). We used two species from the subclass Asterids and order Lamiales as outgroups: Ajuga reptans (NC_023103), and Mimulus guttatus (NC_018041).