Figures & data
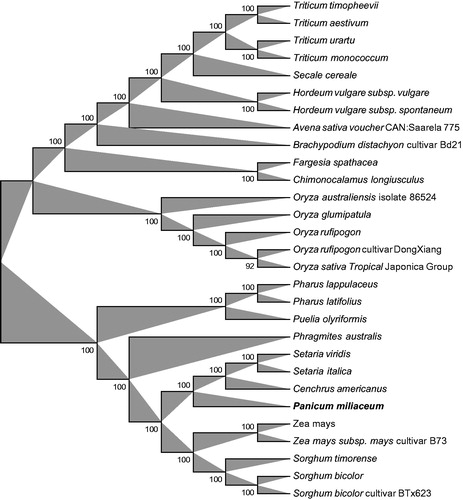
Figure 1. Phylogenetic relationships among 28 whole chloroplast genomes of Gramineae family. The 29 species can be divided into two independent clades. Bootstrap support values are given at the nodes. Chloroplast genome accession number used in this phylogeny analysis: Triticum timopheevii: NC024764; Triticum aestivum: KC912694; Triticum urartu: NC021762; Triticum monococcum: NC021760; Secale cereale: NC021761; Hordeum vulgare subsp. Vulgare: KC912689; Hordeum vulgare subsp. spontaneum: KC912688; Avena sativa voucher CAN:Saarela 775: NC027468; Brachypodium distachyon cultivar Bd21: EU325680; Fargesia spathacea: NC024716; Chimonocalamus longiusculus: NC024714; Oryza australiensis isolate 86524: NC024608; Oryza glumipatula: KM881640; Oryza rufipogon: NC017835; Oryza rufipogon cultivar DongXiang: KF562709; Oryza sativa Tropical Japonica Group: KT289404; Pharus lappulaceus: NC023245; Pharus latifolius: NC021372; Puelia olyriformis: NC023449; Phragmites australis: NC022958; Setaria viridis: NC028075; Setaria italica: KJ001642; Cenchrus americanus: KJ490012; Zea mays: NC001666; Zea mays subsp. mays cultivar B73: KF241981; Sorghum timorense: NC023800; Sorghum bicolor: NC008602; Sorghum bicolor cultivar BTx623: EF115542. Bold and italic were the sample as Panicum miliaceum in this study.