Figures & data
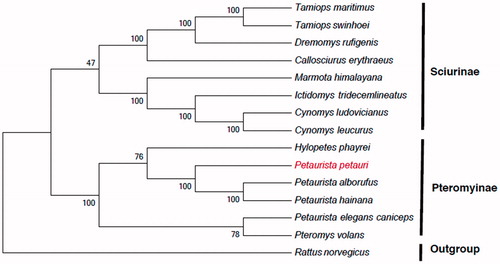
Figure 1. Maximum-likelihood tree inferred from the nucleotide sequence of whole mitogenome. The numbers above branches specify bootstrap percentages (1000 replicates). GenBank accession numbers for the published sequences are H. phayrei (NC026443); P. volans (JQ230001); P. alborufus (NC023922); P. hainana (JX572159); P. elegans caniceps (KU579289); C. erythraeus (NC025550); C. leucurus (NC026705); C. ludovicianuse (NC026706); D. rufigenis (NC026442); I. tridecemlineatus (NC027278); M. himalayana (NC018367); T. maritimus (NC029325); T. swinhoei (NC026875); R. norvegicus (KF011917).