Figures & data
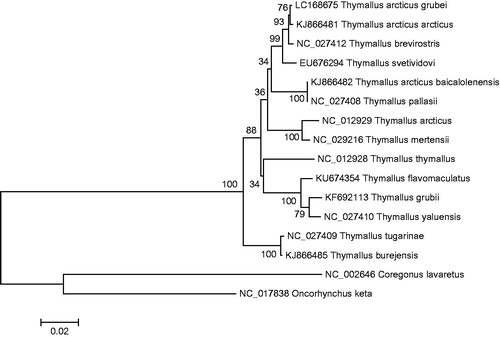
Figure 1. A phylogenetic tree constructed based on the comparison of complete mitochondrial genome sequences of the grayling, T. arcticus grubei and other thirteen species of Thymallus genus. They are T. thymallus (European grayling), T. pallasii (East Siberian grayling), T. arcticus (Arctic grayling), T. tugarinae (Lower Amur grayling), T. burejensis (Bureya grayling), T. grubii (Amur grayling), T. arcticus arcticus (Arctic grayling), T. brevirostris (Mongolian grayling), T. yaluensis (Yalu grayling), T. mertensii (Kamchatka grayling), T. flavomaculatus (Yellow-spotted grayling), T. baicalolenensis (Baikal black grayling), and T. svetividovi (Upper Yenisei grayling). Oncorhynchus keta and Coregonus lavaretus are used as outgroup. Genbank accession numbers for all sequences are listed in the figure. The numbers at the nodes are bootstrap percent probability values based on 1000 replications.