Figures & data
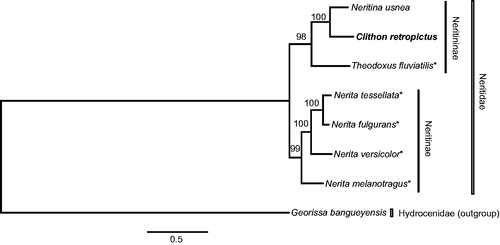
Figure 1. Maximum likelihood phylogeny of Neritidae based on mitochondrial DNA sequences of 13 protein-coding and two ribosomal RNA genes from Clithon retropictus (bold, this study; LC127067), Neritina usnea (KU342665), Theodoxus fluviatilis (KU342667), Nerita fulgurans (KF728888), Nerita melanotragus (GU810158), Nerita tessellata (KF728889), Nerita versicolor (KF728890) and Georissa bangueyensis (outgroup; KU342664). Asterisks denote species for which a near-complete mitogenome is available. Sequences were aligned separately for each gene using Translator X (for coding genes; Abascal et al. Citation2010) and MAFFT v7 (rRNA genes; Katoh & Standley Citation2013) with default parameters. Ambiguously aligned positions were removed using Gblocks v.0.91b (Castresana Citation2000) with all options for a less stringent selection. Tree reconstruction was performed in RAxML v.7.4.2 (Stamatakis Citation2006) using GTR + G model; nodal support estimated by 1000 thorough bootstrap replicates. Scale bar represents branch length (substitutions/site). Three limnic taxa of subfamily Neritininae (Clithon, Neritina and Theodoxus) were recovered as a robust clade (bootstrap probability: 98%) and sister to the also monophyletic Neritinae (Nerita; 99%).