Figures & data
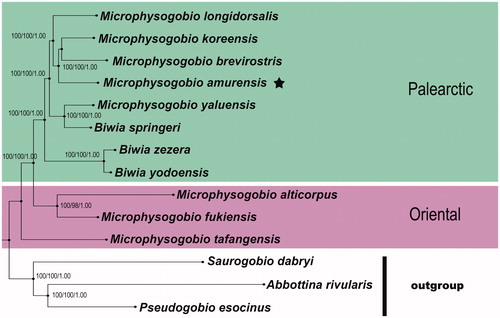
Figure 1. Phylogenetic relationships among M. amurensis and other Asian dwarf gudgeons with complete mitogenome sequences on GenBank were inferred using the Maximum-Likelihood (ML), Neighbour-Joining (NJ), and Bayesian (BA) phylogenetic analyses, respectively. Numbers above branches are bootstrap support of the ML, NJ analysis higher than 95 and posterior probabilities of the Bayesian analysis higher than 95%, for the combined analysis. The results for the entire mitogenomes gave with three methods very similar tree topologies. The gene’s accession numbers for tree construction are listed as follows: M. tafangensis (KF857260.1), M. alticorpus (KC762939.1), M. fukiensis (KJ933414.1), M. brevirostris (KF319122.1), M. yaluensis (KR075133.1), M. koreensis (JX179157.1), M. longidorsalis (AP011394.1), Biwia zezera (AB250107.1), B. yodoensis (AB250108.1), B. springeri (AP011360.1), Abbottina rivularis (AP011257.1), Saurogobio dabryi (KF534790.1), Pseudogobio esocinus (AP009310.1). Clades with taxa inhabited different biogeographic realms were marked by various shading/colours (Oriental: the Oriental species; Palearctic: the Palearctic species). The internal placements of Biwia taxa from Japan and Korea within the genus Microphysogobio implied that their independent generic positions were not supported herein, and they probably could be placed in a single genus. If the priority status of Microphysogobio vs. Biwia is confirmed, the latter being a senior synonym.