Figures & data
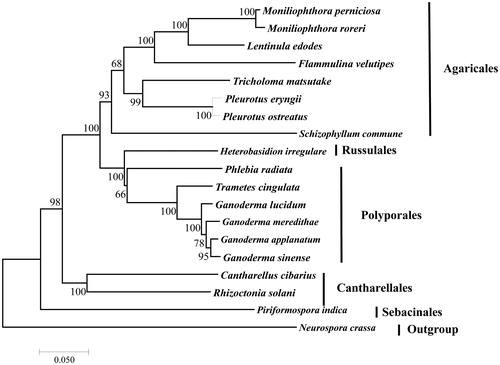
Figure 1. Phylogenetic analysis of 18 species of Agaricomycotina (including L. edodes) conducted based on the NJ method implemented in MEGA 7.0 (Kumar et al. Citation2016). A total of 11 amino acid sequences were used, including atp8, atp9, cob, cox1, cox2, cox3, nad1, nad3, nad4L, nad5, and nad6. The concatenated sequences were aligned using Clustal (Thompson et al. Citation2010). All the sequences could be currently available in the GenBank database: Cantharellus cibarius (NC_020368), Flammulina velutipes (NC_021373), Ganoderma applanatum (NC_027188), Ganoderma lucidum (NC_021750), Ganoderma meredithae (NC_026782), Ganoderma sinense (NC_022933), Heterobasidion irregulare (NC_024555), Moniliophthora perniciosa (NC_005927), Moniliophthora roreri (NC_015400), Phlebia radiata (NC_020148), Pleurotus eryngii (KX827267), Pleurotus ostreatus (NC_009905), Rhizoctonia solani (HF546977), Schizophyllum commune (NC_003049), Serendipita indica (FQ859090), Trametes cingulata (NC_013933), and Tricholoma matsutake (NC_028135). Neurospora crassa (NC_026614) was served as an outgroup. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches.