Figures & data
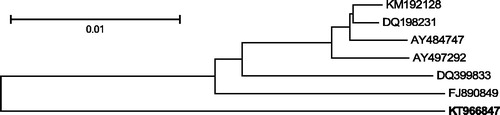
Figure 1. Comparative analysis of the announced mitogenome. The following six complete F mitochondrial genomes, most similar (more than 94% sequence similarity) to the announced F genome of M. chilensis (KT966847, in bold) have been downloaded from GenBank: KM192128 and DQ198231 M. trossulus from the Baltic Sea (Zbawicka et al. Citation2007, Citation2014), AY484747 M. edulis from the Atlantic (Boore et al. Citation2004), AY497292 M. galloprovincialis from the Atlantic (Mizi et al. Citation2005), DQ399833 M. galloprovincialis from the Black Sea (Venetis et al. Citation2007), FJ890849 M. galloprovincialis from the Mediterranean Sea (Burzyński & Smietanka Citation2009). The sequences were aligned using MUSCLE (Edgar Citation2004). All alignment positions containing gaps and missing data were eliminated. There were a total of 16,247 positions in the final data set. The tree was inferred using the Neighbour-Joining method (Saitou & Nei Citation1987). The distances between sequences were computed as the number of base differences per site. The tree is drawn to scale, with branch lengths in the same units. The optimal tree, with the sum of branch lengths = 0.08 is shown. Bootstrap support (1000 replicates, Felsenstein Citation1985) was greater than 95% for all bipartitions. All analyses were conducted in MEGA6 (Tamura et al. Citation2013).