Figures & data
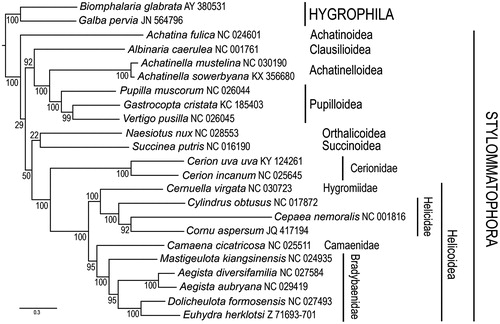
Figure 1. Placement of Cerion uva uva among Stylommatophoran snails. Amino-acid sequences of protein-coding genes were individually aligned using the L-INS-i option with default parameters of the MAFFT v. 7 aligner (Katoh & Stanley Citation2013). Nucleotide alignments for individual protein-coding genes were obtained according to their amino-acid alignments using PAL2NAL (Suyama et al. Citation2006). Ribosomal genes were individually aligned using MAFFT (Q-INS-i option) (Katoh & Toh Citation2008). Individual nucleotide gene alignments were filtered using Gblocks (Talavera & Castresana Citation2007) with default parameters, allowing gaps in all positions. These were subsequently concatenated, leading to alignments with 9785 nucleotide positions for protein-coding genes and 1282 positions for the rRNA genes. Maximum likelihood analyses were performed using RAxML v. 8 (Stamatakis Citation2014). The general time reversible (GTR) model of nucleotide evolution was used. Maximum likelihood analyses consisted of 1000 independent tree searches and bootstrap runs. All analyses were run on the Smithsonian Institution’s high-performance computing cluster (SI/HPC). The resulting trees show similar relationships to previous studies (Gonzalez et al. Citation2016).