Figures & data
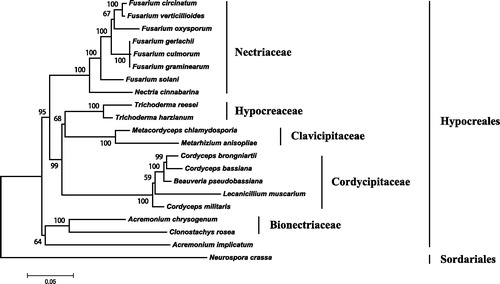
Figure 1. Phylogenetic relationship of 20 taxa of Hypocreales (Ascomycota) determined by neighbor-joining analysis based on concatenated sequences of 15 translated mitochondrial proteins. The 15 proteins included subunits of the respiratory chain complexes (cob, cox1, cox2, cox3), ATPase subunits (atp6, atp8, and atp9), NADH: quinone reductase subunits (nad1, nad2, nad3, nad4, nad4L, nad5, nad6), and ribosomal protein S3 (rps3). The concatenated sequences were aligned using MAFFT. The following 19 mitogenomes were used in this analysis: Acremonium chrysogenum (NC_023268), A. implicatum (NC_026534), Beauveria pseudobassiana (NC_022708), Cordyceps bassiana (NC_010652), C. brongniartii (NC_011194), C. militaris (NC_022834), Fusarium circinatum (NC_022681), F. culmorum (NC_026993), F. gerlachii (NC_025928), F. graminearum (NC_009493), F. oxysporum (NC_017930), F. solani (NC_016680), F. verticillioides (NC_016687), Lecanicillium muscarium (NC_004514), Metacordyceps chlamydosporia (NC_022835), Metarhizium anisopliae (NC_008068), Nectria cinnabarina (KT731105), Trichoderma harzianum (KR952346) and T. reesei (NC_003388). Neurospora crassa (NC_026614) was served as an outgroup. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches.