Figures & data
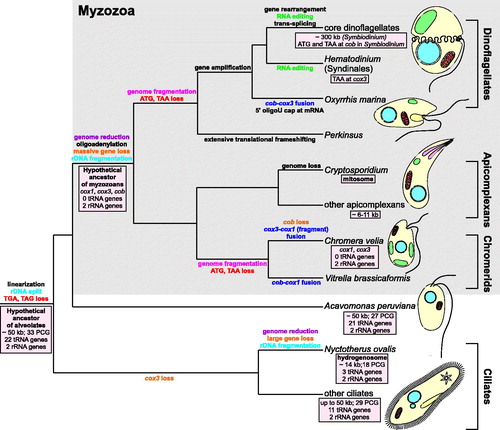
Figure 1. Evolution of mitochondrial genomes in alveolates. The ancestral genome of the superphylum Alveolata was probably about 50 kb in size, such as the mitogenomes of extant ciliates and Acavomonas peruviana, and encoded about 60 genes (Janouskovec et al. Citation2013). Since alveolates diverged about 850 million years ago (Berney & Pawlowski Citation2006), its mitogenome was subjected to various, often convergent, modifications and transformations as subsequent lineages of the superphylum radiated. Initially, the alveolate mitogenome was linearized from a circular form, lost two stop codons (TGA, TAG) and the genes coding for large and small ribosomal subunits were split into two separately encoded fragments. The most spectacular genome reduction occurred in the common ancestor of myzozoans, i.e. apicomplexans and dinoflagellates, and related lineages (e.g. chromerids), after their divergence from Acavomonas peruviana (Janouskovec et al. Citation2013). Extensive gene loss and gene transfer to the nuclear genome resulted in their extremely small mitogenomes containing only three protein-coding genes (cox1, cox3, and cob), sometimes fused, and two rRNA genes, which were subjected independently to further fragmentation in chromerids and the ancestor of dinoflagellates and perkinsids. This extremely small set of genes was even further reduced in Chromera velia as the chromerid completely lost the cob gene (Obornik & Lukes Citation2015). The myzozoan mitogenomes also got rid of about 20 tRNA genes present in the ancestral alveolate genome. At that time, oligoadenylation of transcripts probably evolved. A similar substantial genome reduction also occurred in the anaerobic ciliate Nyctotherus ovalis, whose mitochondrion was transformed into a hydrogenosome (de Graaf et al. Citation2011). This organelle produces hydrogen, which is utilized by methane-producing archaea living together with Nyctotherus as endosymbionts in the hindgut of cockroaches (de Graaf et al. Citation2011). The genome reduction went to extremes in the respiratory and intestinal parasite Cryptosporidium that completely lost its mitochondrial genetic material and transformed its mitochondrion into a mitosome, probably involved in Fe-S cluster assembly (Keithly et al. Citation2005). In dinoflagellates, genes were amplified to numerous copies, which resulted in an increase in their genome size amounting e.g. to ∼300 kb in Symbiodinium (Shoguchi et al. Citation2015). In these protists and sister lineages, various interesting molecular mechanism evolved, such as: translational frameshifting, the addition of 8–9 uridine caps at 5′ end of mRNAs, trans-splicing, and RNA editing (Flegontov & Lukeš Citation2012). The latter evolved, probably independently, in core dinoflagellates and Syndinales because editing sites are not conserved between these groups, which are separated by lineages without RNA editing (not shown in the figure; Flegontov & Lukeš Citation2012). It is assumed that universal start (ATG) and stop (TAA) codons, still present in apicomplexans, were independently lost in chromerids and the perkinsid-dinoflagellate branch. Given this model, the presence of TAA at the cox3 gene in Hematodinium (Jackson et al. Citation2012) as well as ATG and TAA at cob in Symbiodinium (Shoguchi et al. Citation2015) implies that these codons may have originated de novo. Alternatively, these codons might represent an ancestral state and many alveolates (Perkinsus, Oxyrrhis, and other dinoflagellates) lost these codons independently. PCG: protein-coding genes.