Figures & data
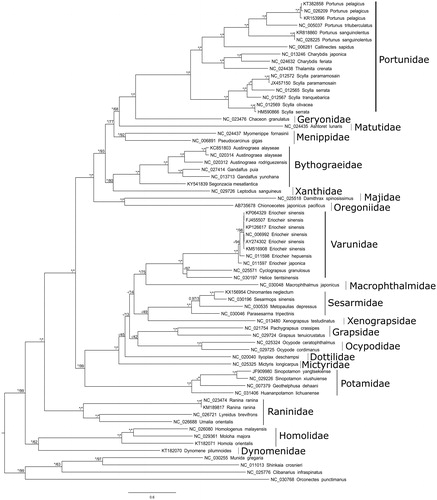
Figure 1. RaxML topology of Brachyura phylogenetic relationships based on the alignment of the concatenated 13 mitochondrial coding genes for 63 Brachyuran and 4 non-Brachyuran decapods. The model GTR + G + I was used in both maximum likelihood and Bayesian analysis. 500 rapid bootstrap resampling were set for RaxML analysis. Convergence of the 2 run Bayesian analysis was verified by the ESS values >200 with Tracer V.1.6. Node values correspond to Bayesian posterior probabilities/bootstrap. Asterisks indicate nodes with complete support in the considered analysis. A hyphen denotes a node not recovered in the considered analysis. All non-redundant available complete mitogenomes were selected and a primary RaxML analysis with a concatenated dataset was performed. The Leucosiidae Pyrhila pisum (NC_030047) generates a long branch and was thus excluded. Each gene was then separately analysed to detect potential long-branch artefacts. This analysis conducted to exclude the Cytb and ND6 genes of Metopaulias depressus (NC_030535). The dataset was completed with Sinopotamon yangtsekiense (JF909980) for which the sequences of ND1 and ND2 are lacking.