Figures & data
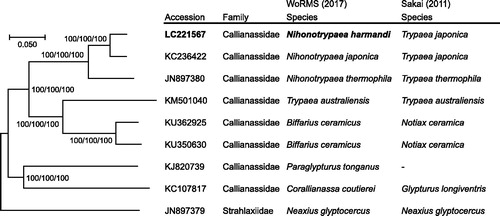
Figure 1. Phylogenetic relationships of the callianassid ghost shrimps inferred from a combined analysis of 13 CDSs and 2 rRNAs using NJ (K2P model), ML (GTR + I + G model) and MP methods each with 1,000 replicates of bootstrap. There were a total of 12,675 positions in the dataset. The tree shown is an NJ tree, and the ML and MP trees were the same topology. Numbers above nodes are bootstrap support values (NJ/ML/MP). Neaxius glyptocercus (accession number: JN897379) was used as outgroup. Classifications of species proposed by WoRMS Editorial Board (Citation2017) and Sakai (Citation2011) are listed. The original species name (source organism in GenBank) of the sequences used for tree reconstructions are listed as follows: KC236422 (Nihonotrypaea japonica), JN897380 (Nihonotrypaea thermophile), KM501040 (Trypaea australiensis), KU362925 (Callianassa ceramica), KU350630 (Callianassa ceramica), KJ820739 (Paraglypturus tonganus), KC107817 (Corallianassa coutierei), JN897379 (Neaxius glyptocercus).