Figures & data
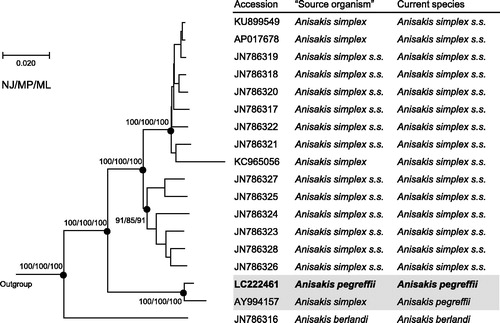
Figure 1. Phylogenetic relationships of the three sibling species of the A. simplex species complex (A. simplex sensu lato) inferred from the complete mitogenome analysis using NJ (K2P model), MP and ML (GTR + I + G model) methods each with 1000 replicates of bootstrap (100 replicates for ML). There were a total of 13,457 positions in the dataset. The tree shown is an NJ tree, and the ML and MP trees were almost the same topology. Bootstrap support values are shown for the major nodes. The three species, Pseudoterranova decipiens sensu lato Germany, Toxocara canis, Ascaris suum (accession numbers: KU558723, AP017701, X54253, respectively) were used as outgroup. ‘Source organism’ is that in GenBank.