Figures & data
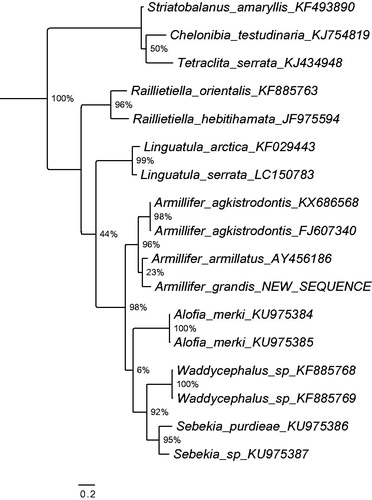
Figure 1. Maximum likelihood tree illustrating the phylogenetic position of the newly sequenced Armillifer grandis gene sequence among a subset of pentastomid species. Cytochrome oxidase I sequences were aligned using MAFFT 7.271 and highly divergent or poorly aligned regions were removed with Gblocks 0.91b (Castresana Citation2000) allowing for gap positions and smaller blocks. Trees were calculated using PhyML 3.1 (Guindon et al. Citation2010) with 12 rate categories, optimized equilibrium frequencies, GTR model of sequence evolution and combined heuristics (Nearest Neighbor Interchange and Subtree Pruning and Rerafting). Branch support was calculated using approximate likelihood ratio tests as implemented in PhyML.