Figures & data
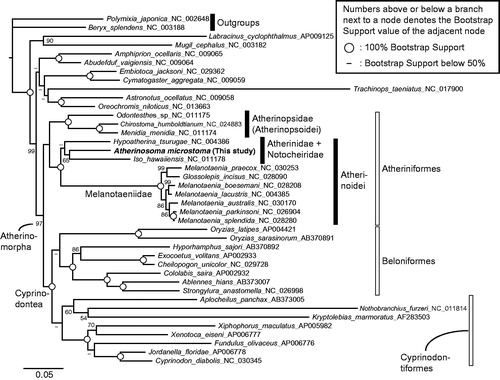
Figure 1. A maximum likelihood (ML) molecular phylogenetic tree, inferred using the program RAxML (Stamatakis Citation2014). Phylogenetic analyses were conducted on a data matrix (11,082 positions) including all the concatenated nucleotide sequences of the mitogenomes except the third codon positions. The ND6 gene was excluded from the analysis. Gene sequences were aligned individually using the online version of MAFFT under default settings (http://mafft.cbrc.jp/alignment/server/; Katoh & Standley Citation2013). Aligned sequences were individually edited using the online version of GBlocks using the least stringent settings (http://molevol.cmima.csic.es/castresana/Gblocks_server.html; Castresana, Citation2000). Data partitions were determined using the program PartitionFinder ver. 2 (Lanfear et al. Citation2017). Partitioned ML analyses were performed with RAxML-GUI ver. 1-5b1 (Silvestro & Michalak Citation2012), with the GTR + Γ + I nucleotide substitution model (Yang Citation1994). The rapid bootstrap analyses were conducted with 1000 replications, with four threads running in parallel.