Figures & data
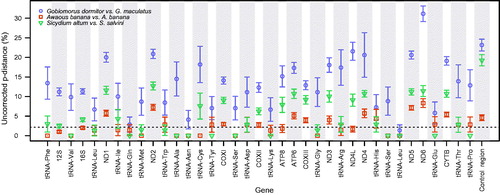
Figure 1. Maximum-likelihood (RAxML) phylogeny using 13 protein-coding mitochondrial genes from a selection of species of Gobiiformes (GenBank accession numbers indicated), and Kurtus gulliveri, which was used as the outgroup. Numbers next to nodes are support values obtained after 500 bootstrap replicates. Sequences from specimens obtained in this study are highlighted in bold.
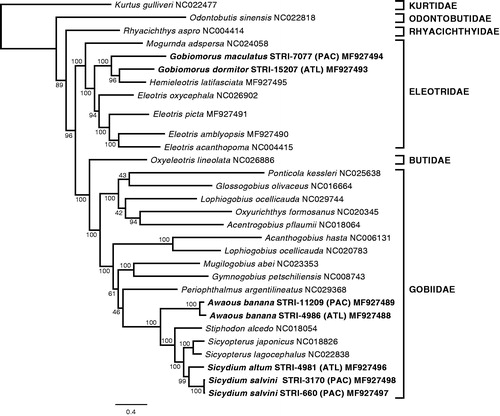
Figure 2. Genetic divergence across all mitochondrial genes between three pairs of species: Gobiomorus dormitor versus G. maculatus, Awaous banana from Atlantic versus A. banana from the Pacific watersheds, and Sicydium salvini versus S. altum. Genes are presented in the same order as they appear in these species mitochondrial genomes. Error bars represent standard errors and the dashed horizontal line indicates the commonly used threshold at 2% genetic divergence.