Figures & data
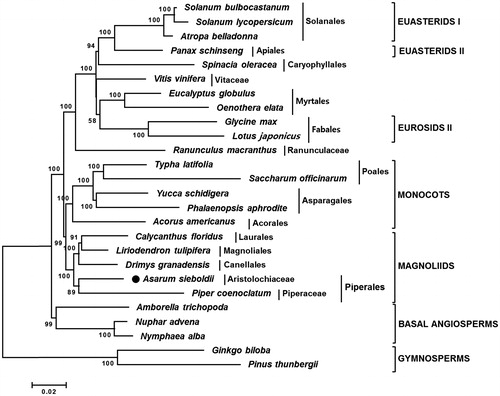
Figure 1. Neighbor-joining (NJ) tree based on the chloroplast protein-coding genes of 26 taxa including A. sieboldii. Sequences of 65 chloroplast protein-coding gene from 26 taxa were aligned using MAFFT (http://mafft.cbrc.jp/alignment/server/index.html) and used to generate NJ phylogenetic tree by MEGA 6.0 (Tamura et al. Citation2013). The numbers in the nodes indicated the bootstrap support values (>50%) from 1000 replicates. Chloroplast genome sequences used for this tree are: Acorus americanus, NC_010093; Amborella trichopoda, NC_005086; A. sieboldii, MG551543; Atropa belladonna, NC_004561; Calycanthus floridus, NC_004993; Drimys granatensis, DQ887676; Eucalyptus globulus, NC_008115; Ginkgo biloba, NC_016986 (outgroup); Glycine max, NC_007942; Liriodendron tulipifera, NC_008326; Lotus japonicus, NC_002694; Nuphar advena, NC_008788; Nymphaea alba, NC_006050; Oenothera elata, NC_002693; Panax schinseng, NC_006290; Phalaenopsis aphrodite, NC_007499; Pinus thunbergii, NC_001631 (outgroup); Piper coenoclatum, DQ887677; Ranunculus macranthus, NC_008796; Saccharum officinarum, NC_006084; Solanum bulbocastanum, NC_007943; Solanum lycopersicum, DQ347959; Spinacia oleracea, NC_002202; Typha latifolia, NC_013823; Vitis vinifera, NC_007957; Yucca schidigera, NC_032714.