Figures & data
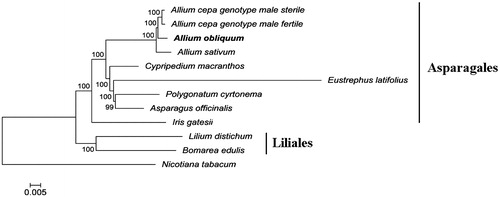
Figure 1. Phylogenetic tree inferred by maximum-likelihood using 82 protein-coding gene sequences of 10 species including seven species from the Asparagales order: Allium cepa (genotype male sterile KF728079 and genotype male fertile NC_024813), Allium sativum (NC_031829), Eustrephus latifolius (KM233639), Polygonatum cyrtonema (KT630835), Cypripedium macranthos (KF925434), Asparagus officinalis (NC_034777), Iris gatesii (KM014691); two species from Liliales order: Bomarea edulis (NC_025306), Lilium distichum (NC_029937); and Nicotiana tabacum (NC_001879) as an outgroup. PhyML 3.1 (Guindon et al. Citation2010) was used for the sequence alignment and construction of the tree. Bootstrap support values based on 1000 replicates are displayed on each node.