Figures & data
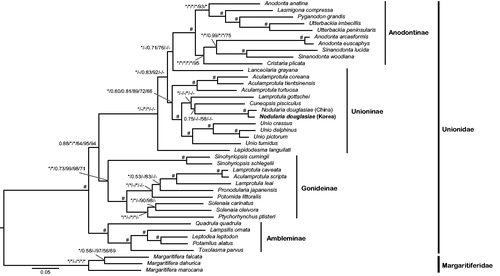
Figure 1. Bayesian inference tree of 38 unionids inferred from 12 PCGs and two rRNA genes, showing phylogenetic relationships among the four unionid subfamilies and phylogenetic position of Nodularia douglasiae. Phylogenetic relationships among 38 unionids based on the three following concatenated data sets: (1) an amino acid sequence alignment set (3259 aa) of 12 PCGs, (2) a full length nucleotide sequence alignment set (11,199 nt) of 12 PCGs and two rRNA genes, and (3) a reduced nucleotide sequence alignment set (7943 nt) of 12 PCGs except for 3rd codon position and two rRNA genes. The branch supporting values on each node are indicated in following order: (1) Bayesian posterior probability based on full length nucleotide sequence alignment, (2) Bayesian posterior probability based on reduced nucleotide sequence alignment except for 3rd codon position, (3) Bayesian posterior probability based on amino acid sequence alignment, (4) bootstrapping values of a maximum-likelihood tree based on full length nucleotide sequence alignment, (5) bootstrapping values of a maximum-likelihood tree based on reduced nucleotide sequence alignment except for 3rd codon position, and (6) bootstrapping values of a maximum-likelihood tree based on amino acid sequence alignment. *Maximum value (BI =1.00 and ML =100). #Case that all six different confidence values on a certain node appear maximums. The Korean N. douglasiae mitochondrial genome obtained from the present study is shown in bold letter on the tree. The three species belonging to the subfamily Margaritiferidae were employed as outgroups in this study.