Figures & data
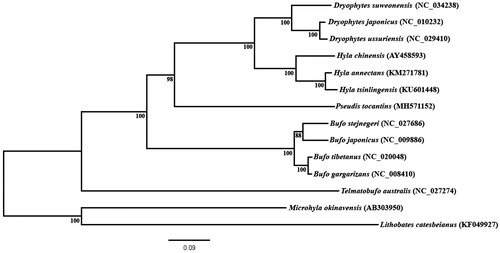
Figure 1. Phylogenetic inference obtained using maximum likelihood based on mitochondrial genomes, excluding control region, of the anuran species Dryophytes japonicus, D. ussuriensis, (as Hyla ussuriensis in Sun et al. Citation2017), D. suweonensis (as H. suweonensis in Lee et al. 2017), Hyla chinensis, H. annectans, Hyla tsinlingensis, Pseudis tocantins, Bufo gargarizans, B. tibetanus (as in Wang et al. 2013), stejnegeri, B. japonicus, Telmatobufo australis, Microhyla okinavensis and Lithobates catesbeianus. M. okinavensis and L. catesbeianus were used as outgroups. The phylogenetic inference was constructed under GTR + G evolutionary model. Bootstrap analysis was performed using 1000 pseudoreplicates for node support (numbers at the nodes).