Figures & data
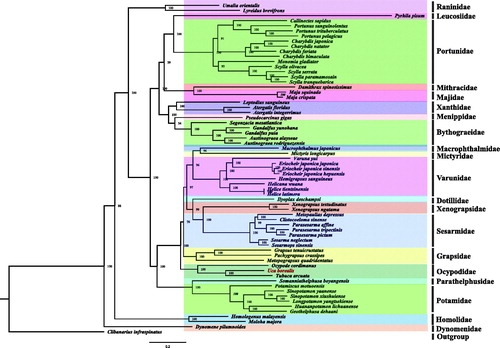
Figure 1. Phylogeny of Brachyura based on nucleotide sequences. The phylogenetic tree was inferred from the nucleotide sequences of 13 mitogenome PCGs using BI and ML methods. Numbers on branches indicate posterior probability (BI). Clibanarius infraspinatus was used as outgroup. The genbank accession numbers for all of the sequences is listed as follows: Atergatis floridus NC_037201.1, Atergatis integerrimus NC_037172.1, Austinograea alayseae NC_020314.1, Austinograea rodriguezensis NC_020312.1, Callinectes sapidus NC_006281.1, Charybdis feriata NC_024632.1, Charybdis japonica NC_013246.1, Charybdis natator NC_036132.1, Clibanarius infraspinatus NC_025776.1, Clistocoeloma sinense NC_033866.1, Dynomene pilumnoides KT182070.1, Eriocheir japonica hepuensis NC_011598.1, Eriocheir japonica japonica NC_011597.1, Eriocheir japonica sinensis NC_006992.1, Gandalfus puia NC_027414.1, Gandalfus yunohana NC_013713.1, Geothelphusa dehaani NC_007379.1, Grapsus tenuicrustatus NC_029724.1, Harybdis bimaculata MG489891.1, Helicana wuana NC_034995.1, Helice latimera NC_033865.1, Helice tientsinensis NC_030197.1, Hemigrapsus sanguineus NC_035307.1, Homologenus malayensis NC_026080.1, Huananpotamon lichuanense NC_031406.1, Ilyoplax deschampsi NC_020040.1, Leptodius sanguineus NC_029726.1, Longpotamon yangtsekiense NC_036946.1, Lyreidus brevifrons NC_026721.1, Macrophthalmus japonicus NC_030048.1, Maguimithrax spinosissimus NC_025518.1, Maja crispata NC_035424.1, Maja squinado NC_035425.1, Metopaulias depressus NC_030535.1, Metopograpsus quadridentatus MH310445, Mictyris longicarpus NC_025325.1, Moloha majora NC_029361.1, Monomia gladiator NC_037173.1, Ocypode cordimanus NC_029725.1, Ortunus pelagicus KR153996.1, Otamiscus motuoensis KY285013.1, Pachygrapsus crassipes NC_021754.1, Parasesarma affine MH310444, Parasesarma pictum MG580780, Parasesarma tripectinis NC_030046.2, Portunus pelagicus NC_026209.1, Portunus sanguinolentus NC_028225.1, Portunus trituberculatus NC_005037.1, Pseudocarcinus gigas NC_006891.1, Pyrhila pisum NC_030047.1, Scylla olivacea NC_012569.1, Scylla paramamosain NC_012572.1, Scylla serrata NC_012565.1, Scylla tranquebarica NC_012567.1, Segonzacia mesatlantica NC_035300.1, Sesarma neglectum NC_031851.1, Sesarmops sinensis NC_030196.1, Sinopotamon xiushuiense NC_029226.1, Sinopotamon yaanense NC_036947.1, Somanniathelphusa boyangensis NC_032044.1, Tubuca arcuata KX911977.1, Umalia orientalis NC_026688.1, Varuna yui NC_037155.1, Xenograpsus ngatama NC_035951.1, Xenograpsus testudinatus NC_013480.1.