Figures & data
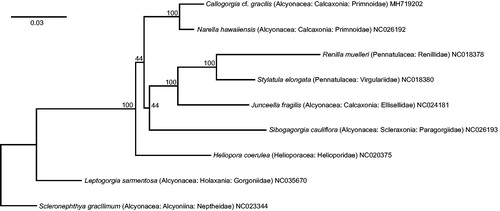
Figure 1. Maximum-likelihood, phylogenetic tree of the complete mitogenomes of Callogorgia cf. gracilis (this study) and eight representative octocorals (taxonomic position and GenBank accession numbers in tip labels). In Geneious R10.2.6, complete mitogenomes were aligned with default MUSCLE parameters; the resulting alignment was used to construct the phylogenetic tree in RAxML 8.2.11 with the following changes to the default settings: bootstrap replicates =1000, algorithm = rapid bootstrapping and search for best-scoring ML tree, outgroup = NC023344, nucleotide model = GTR CAT I. Bootstrap values are reported at the nodes. See figshare (DOI: 10.6084/m9.figshare.6998459) for extended methods.