Figures & data
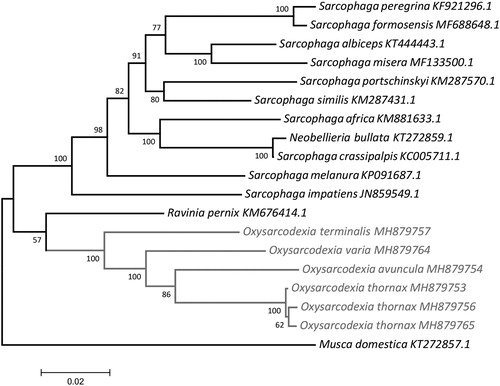
Figure 1. Molecular phylogenetic inferences of Oxysarcodexia mitogenomes. The phylogenetic analysis was based on the maximum likelihood (ML) with the nearest-neighbor-interchange heuristic method using MEGA 7 software (Kumar et al. Citation2016). The D-loop region was excluded from this analysis due to its high degree of variability (Gonder et al. Citation2007). The tree showed a clearly separated branch for the species and suggests that the Oxysarcodexia clade is monophyletic and well-supported by the branches and is closer to the genus Ravina. The individual differences among O. thornax specimens may be due to the geographical separation of the populations, as the individuals MH879756 and MH879765, collected from locations separated by approximately 100 km, showed higher similarity to each other in comparison with MH879753, which was collected approximately 1300 and 1400 km away from MH879765 and MH879756, respectively. The Oxysarcodexia specimens are highlighted in grey. The mitogenomes for comparison were obtained from the NCBI database: Musca domestica (KT272857.1) – used as outgroup, Sarcophaga impatiens (JN859549.1), Sarcophaga melanura (KP091687.1), Neobellieria bullata (KT272859.1), Sarcophaga crassipalpis (KC005711.1), Sarcophaga peregrina (KF921296.1), Sarcophaga formosensis (MF688648.1), Sarcophaga africa (KM881633.1), Sarcophaga portschinskyi (KM287570.1), Sarcophaga similis (KM287431.1), Sarcophaga albiceps (KT444443.1), Sarcophaga misera (MF133500.1), Ravinia pernix (KM676414.1).