Figures & data
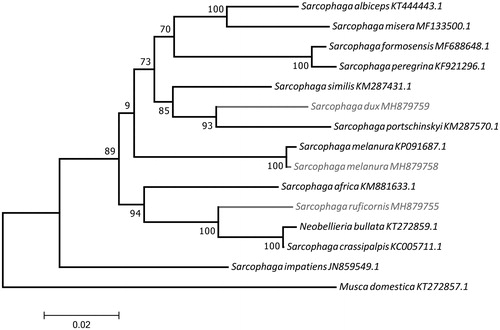
Figure 1. Molecular phylogenetic inferences of Sarcophaga mitogenomes. The phylogenetic analyses were made using the MEGA 7 software (Kumar et al.Citation2016) using the maximum likelihood model with the Nearest-Neighbour-Interchange heuristic method. The D-loop region was excluded from this analysis due to its high degree of variability (Gonder et al.,Citation2007). The tree showed a well-structured separation of the novel species. S. melanura (collected in Lisbon, Portugal) showed few differences in comparison with the species from NCBI collected in China (KP091687.1). S. dux (Aroeira, Portugal) is more related to S. portschinskyi (China). On the other hand, S. ruficornis (Brazil) clustered with the branch including S. crassipalpis and N. bullata (both from the United States). The novel mtDNA published herein are highlighted in grey. The mitogenomes for comparison were obtained from the NCBI database: Musca domestica (KT272857.1) – used as outgroup, Sarcophaga impatiens (JN859549.1), Sarcophaga melanura (KP091687.1), Neobellieria bullata (KT272859.1), Sarcophaga crassipalpis (KC005711.1), Sarcophaga peregrina (KF921296.1), Sarcophaga formosensis (MF688648.1), Sarcophaga africa (KM881633.1), Sarcophaga portschinskyi (KM287570.1), Sarcophaga similis (KM287431.1), Sarcophaga albiceps (KT444443.1), Sarcophaga misera (MF133500.1).