Figures & data
Figure 1. Top: Origin of the re-arranged tRNA cluster Gln, Ile, Met. Most Gobiidae carry the arrangement Ile, Gln, Met without spacers. Benthophilinae however carry the arrangement Gln, Ile, Met, and feature variable length spacers between the genes. Bottom: Alignment of Benthophilinae tRNA sequences. Spacer sequences vary between species, but display more similarities among Neogobius species (Neogobius melanostomus, Neogobius fluviatilis) and Ponticola species (Proterorhinus semilunaris, Babka gymontrachelus, Ponticola kessleri), respectively.
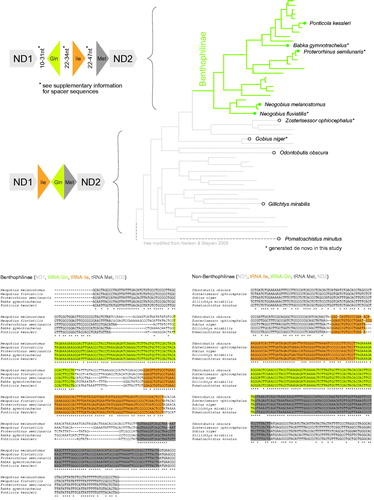
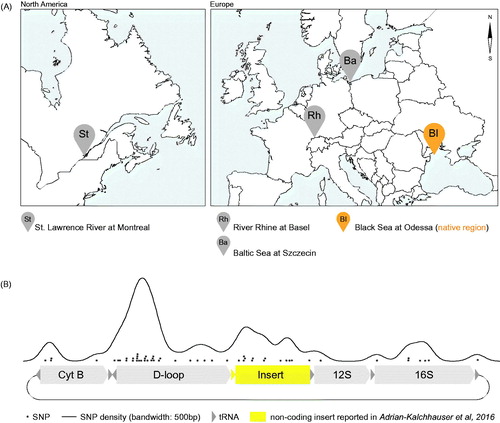
Figure 2. Global occurrence of the novel sequence insertion. (A) Map of sampling sites in North America (St. Lawrence river) and Europe (Rhine river, Baltic Sea, and native region Black Sea). (B) Density of single nucleotide polymorphisms observed in the novel round goby non-coding insert, the adjacent D-loop, and adjacent expressed coding and non-coding sequences.