Figures & data
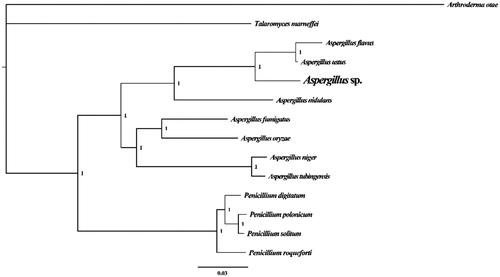
Figure 1. Molecular phylogenies of 13 species based on Bayesian inference analysis of the combined mitochondrial gene set (14 core protein-coding genes + 2 rRNA genes). Node support values are Bayesian posterior probabilities. Mitogenome accession numbers used in this phylogeny analysis: Aspergillus flavus (NC_026920), Aspergillus fumigatus (NC_017016), Aspergillus nidulans (NC_017896), Aspergillus niger (NC_007445), Aspergillus oryzae (NC_018100), Aspergillus tubingensis (NC_007597), Aspergillus ustus (NC_025570), Penicillium digitatum (NC_015080), Penicillium polonicum (NC_030172), Penicillium roqueforti (NC_027416), Penicillium solitum (NC_016187), Talaromyces marneffei (NC_005256), Arthroderma otae (NC_012832).