Figures & data
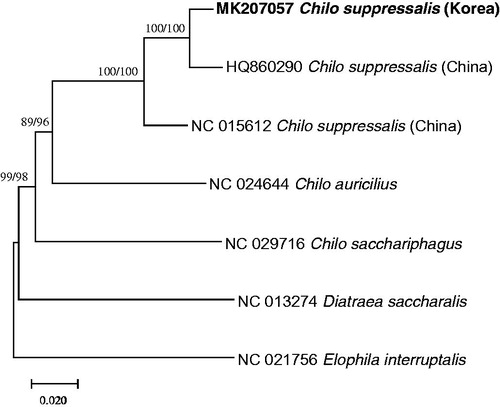
Figure 1. maximum likelihood (bootstrap repeat is 1,000) and neighbor joining (bootstrap repeat is 10,000) phylogenetic trees based on seven complete mitochondrial genomes in Crambidae: Korean C. suppressalis (MK207057, this study), Chinese C. suppressalis (NC_015612 and HQ860290), Chilo sacchariphagus (NC_029716), Chio auricilius (NC_024644), Diatraea saccharalis (NC_013274), and Elophila interruptalis (NC_021756). The numbers above the branches indicate bootstrap support values of maximum likelihood and neighbor joining trees.