Figures & data
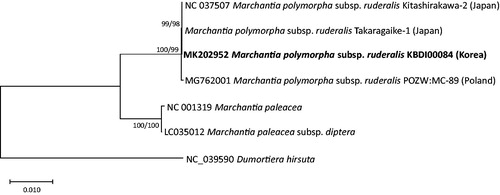
Figure 1. Maximum likelihood (bootstrap repeat is 1000) and neighbor joining (bootstrap repeat is 10,000) phylogenetic trees of Marchantiaceae based on seven complete chloroplast genomes including one outgroup species: M. polymorpha subsp. ruderalis (MK202952 in this study, NC_037507, Takaragaike-1, and MG762001), Marchantia paleacea (NC_001319), Marchantia paleacea subsp. diptera (LC035012), and Dumortiera hirsuta (NC_039590) as an outgroup species. The numbers above branches indicate bootstrap support values of maximum likelihood and neighbor joining phylogenetic trees, respectively.