Figures & data
Figure 1. Sampling localities in this study and phylogenetic relationships of Quasipaa shini. (A) Sampling localities (P1–P8) in this study. See information of populations P1–P8 in . Size of pie charts denoted beside localities is in proportion to number of samples, and different colors represent different clades/subclades. (B) Maximum likelihood tree based on COI gene sequences of haplotypes. Bootstrap supports of major nodes were denoted near them. Insert: the lower and upper frog photos are the phenotype of Clades I and II, respectively. (C) Haplotype network. The circle size is proportional to number of samples. Each subclade was surrounded by a dotted line. One white dot means one mutation step.
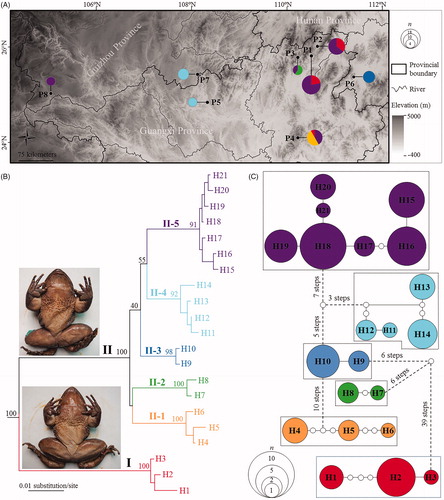
Table 1. Genetic diversity and neutrality tests of Quasipaa shini.
