Figures & data
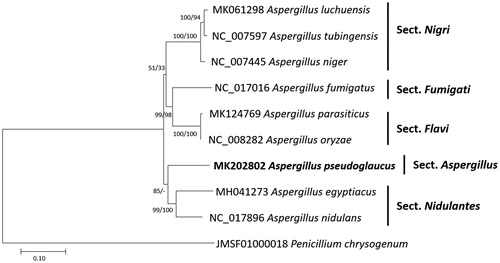
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of nine Aspergillus and one Penicillium mitochondrial genome: Aspergillus pseudoglaucus (MK202802, this study), Aspergillus parasiticus (MK124769), Aspergillus luchuensis (MK061298), Aspergillus egyptiacus (MH041273), Aspergillus tubingensis (NC_007597), Aspergillus nidulans (NC_017896), Aspergillus niger (NC_007445), Aspergillus oryzae (NC_008282), Aspergillus fumigatus (NC_017016), and Penicillium chrysogenum (JMSF01000018). The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees, respectively.