Figures & data
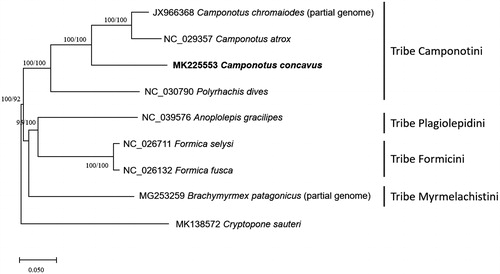
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of all available ants mitochondrial genomes in Formicinae subfamily: Camponotus concavus (This study; MK225553), Camponotus atrox (NC_029357), Camponotus chromaiodes (JX966368; partial genome), Polyrhachis dives (NC_030790), Anoplolepis gracilipes (NC_039576), Formica fusca (NC_026132), Formica selysi (NC_026711), Brachymyrmex patagonicus (MG253259; partial genome), and Cryptopone sauteri (MK138572) as an outgroup species. The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood methods.