Figures & data
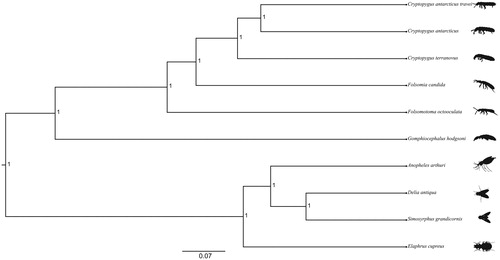
Figure 1. Bayesian phylogenetic tree of Cryptopygus antarcticus travei and nine other species (>69 identity) (five springtail species and four hexapod species). The following species and GenBank accession numbers used for the phylogenetic analysis are as follows: Cryptopygus antarcticus travei (MK433191), Cryptopygus antarcticus (NC_010533), Cryptopygus terranovus (NC_037610), Folsomia candida (KU198392), Folsomotoma octooculata (NC_024155), Gomphiocephalus hodgsoni (NC_005438), Anopheles arthuri (NC_037806), Delia antiqua (NC_028226), Simosyrphus grandicornis (NC_008754), and Elaphrus cupreus (KX087286). All the nodes on the tree are supported by a posterior probability of 1.