Figures & data
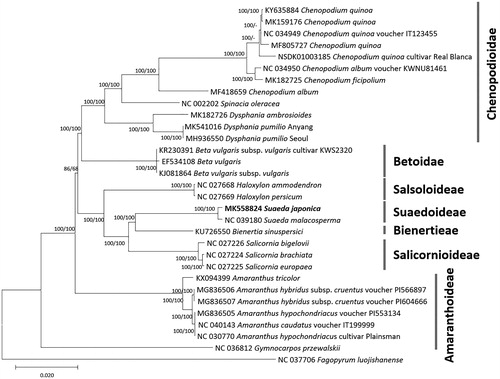
Figure 1. Neighbor joining (bootstrap repeat is 10,000) and maximum likelihood (bootstrap repeat is 1,000) phylogenetic trees of 31 Amaranthaceae complete chloroplast genomes: Suaeda japonica (MK558824 in this study), Suaeda malacosperma (NC_039180), Chenopodium ficifolium (MK182725), Chenopodium quinoa (NC_034949, MK159176, MF805727, KY635884, and NSDK01003185), Chenopodium album (NC_034950 and MF418659), Spinacia oleracea (NC_002202), Dysphania pumilio (MH936550 and MK541016), Dysphania ambrosioides (MK182726), Beta vulgaris (EF534108), Beta vulgaris subsp. vulgaris (KJ081864), Beta vulgaris subsp. vulgaris ‘KWS2320’ (KR230391), Salicornia brachiate (NC_027224), Salicornia europaea (NC_027225), Salicornia bigelovii (NC_027226), Bienertia sinuspersici (KU726550), Haloxylon ammodendron (NC_027668), Haloxylon persicum (NC_027669), Amaranthus tricolor (KX094399), Amaranthus hypochondriacus (NC_030770 and MG836505), Amaranthus caudatus (NC_040143), Amaranthus hybridus subsp. cruentus (MG836506 and MG836507), Gymnocarpos przewalskii (NC_036812), Fagopyrum luojishanense (NC_037706). Phylogenetic tree was drawn based on neighbor joining tree. Right bars indicate each subfamily in the tree. The numbers above branches indicate bootstrap support values of neighbor joining and maximum likelihood phylogenetic trees, respectively.